Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3856-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3856-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 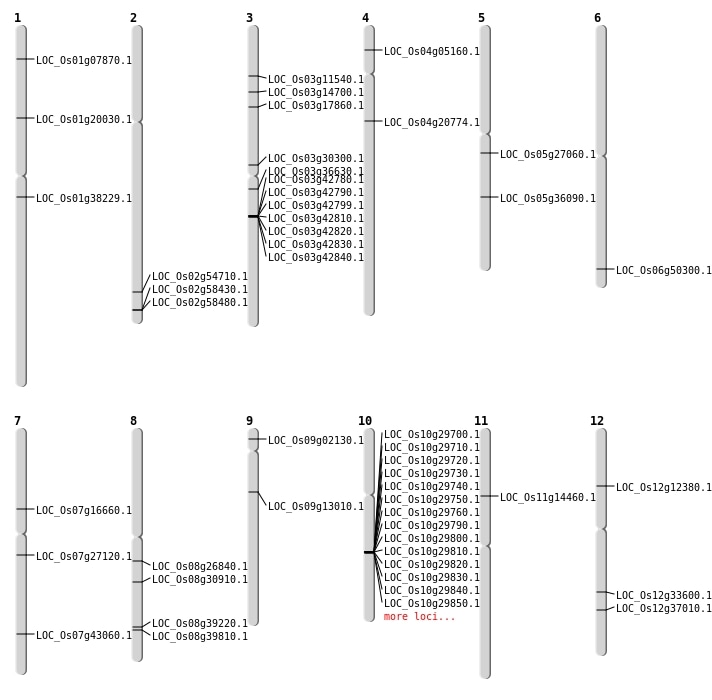
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11379529 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g20030 |
| SBS | Chr1 | 23040427 | C-T | HET | ||
| SBS | Chr1 | 29163891 | A-G | HET | ||
| SBS | Chr1 | 32952566 | T-A | Homo | ||
| SBS | Chr1 | 3807514 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g07870 |
| SBS | Chr11 | 20218643 | G-A | HET | ||
| SBS | Chr11 | 2097945 | A-T | Homo | ||
| SBS | Chr11 | 27274883 | A-G | HET | ||
| SBS | Chr11 | 8121761 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g14460 |
| SBS | Chr12 | 10179501 | C-T | HET | ||
| SBS | Chr12 | 11217375 | A-G | HET | ||
| SBS | Chr12 | 11668697 | T-C | Homo | ||
| SBS | Chr12 | 19270139 | T-A | HET | ||
| SBS | Chr12 | 4268389 | G-A | HET | ||
| SBS | Chr2 | 12540662 | C-T | Homo | ||
| SBS | Chr2 | 2471933 | T-A | HET | ||
| SBS | Chr2 | 31390685 | A-G | HET | ||
| SBS | Chr2 | 32678535 | C-T | HET | ||
| SBS | Chr2 | 35106023 | T-C | HET | ||
| SBS | Chr2 | 35734030 | C-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g58430 |
| SBS | Chr3 | 20053998 | C-T | HET | ||
| SBS | Chr3 | 20756714 | T-C | HET | ||
| SBS | Chr3 | 27460500 | A-T | Homo | ||
| SBS | Chr3 | 33510828 | A-G | HET | ||
| SBS | Chr3 | 35006154 | C-T | HET | ||
| SBS | Chr4 | 1213471 | G-A | HET | ||
| SBS | Chr4 | 16189774 | C-T | HET | ||
| SBS | Chr4 | 19218365 | C-T | HET | ||
| SBS | Chr4 | 8619934 | C-T | HET | ||
| SBS | Chr5 | 15734390 | T-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g27060 |
| SBS | Chr5 | 17164999 | C-A | Homo | ||
| SBS | Chr5 | 24580767 | C-T | Homo | ||
| SBS | Chr5 | 9902966 | A-G | HET | ||
| SBS | Chr6 | 8962767 | G-A | HET | ||
| SBS | Chr7 | 25800684 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g43060 |
| SBS | Chr8 | 17760231 | G-A | HET | ||
| SBS | Chr8 | 19088777 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g30910 |
| SBS | Chr8 | 24775024 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g39220 |
| SBS | Chr8 | 8519497 | A-C | HET | ||
| SBS | Chr9 | 13666408 | A-G | HET | ||
| SBS | Chr9 | 22849588 | T-C | HET | ||
| SBS | Chr9 | 22851547 | G-T | HET |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 873065 | 873067 | 2 | |
| Deletion | Chr8 | 2132607 | 2132608 | 1 | |
| Deletion | Chr9 | 2440769 | 2440771 | 2 | |
| Deletion | Chr4 | 2571328 | 2571330 | 2 | LOC_Os04g05160 |
| Deletion | Chr7 | 4683451 | 4683455 | 4 | |
| Deletion | Chr6 | 4756530 | 4756532 | 2 | |
| Deletion | Chr2 | 7832011 | 7832129 | 118 | |
| Deletion | Chr3 | 7993910 | 7993916 | 6 | LOC_Os03g14700 |
| Deletion | Chr2 | 9518395 | 9518398 | 3 | |
| Deletion | Chr12 | 11979507 | 11979661 | 154 | |
| Deletion | Chr3 | 13106943 | 13107034 | 91 | |
| Deletion | Chr5 | 15399650 | 15399658 | 8 | |
| Deletion | Chr10 | 15441001 | 15860000 | 418999 | 70 |
| Deletion | Chr10 | 15485829 | 15485831 | 2 | |
| Deletion | Chr10 | 21941471 | 21941473 | 2 | |
| Deletion | Chr6 | 22384700 | 22384764 | 64 | |
| Deletion | Chr11 | 22688879 | 22689204 | 325 | |
| Deletion | Chr7 | 22711678 | 22711686 | 8 | |
| Deletion | Chr3 | 23829001 | 23898000 | 68999 | 7 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 21829693 | 21829759 | 67 | |
| Insertion | Chr12 | 22687920 | 22687920 | 1 | LOC_Os12g37010 |
| Insertion | Chr5 | 194174 | 194177 | 4 |
Inversions: 29
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 805729 | 805913 | LOC_Os09g02130 |
| Inversion | Chr5 | 1472128 | 1472410 | LOC_Os05g03480 |
| Inversion | Chr12 | 2245126 | 2245301 | |
| Inversion | Chr8 | 4046517 | 4046883 | |
| Inversion | Chr3 | 5982840 | 5982955 | LOC_Os03g11540 |
| Inversion | Chr12 | 6818020 | 6818246 | LOC_Os12g12380 |
| Inversion | Chr3 | 9954000 | 9954197 | LOC_Os03g17860 |
| Inversion | Chr4 | 11659160 | 11659385 | LOC_Os04g20774 |
| Inversion | Chr11 | 12290835 | 12291265 | |
| Inversion | Chr9 | 12388157 | 12388501 | |
| Inversion | Chr8 | 12797072 | 12797284 | |
| Inversion | Chr10 | 15446044 | 16056326 | LOC_Os10g30826 |
| Inversion | Chr7 | 15729883 | 15730249 | LOC_Os07g27120 |
| Inversion | Chr10 | 15863061 | 16056725 | LOC_Os10g30826 |
| Inversion | Chr8 | 16322024 | 16322296 | LOC_Os08g26840 |
| Inversion | Chr3 | 17288197 | 17288325 | LOC_Os03g30300 |
| Inversion | Chr12 | 20291392 | 20291519 | LOC_Os12g33600 |
| Inversion | Chr3 | 20317775 | 20318117 | LOC_Os03g36630 |
| Inversion | Chr12 | 20535788 | 20535978 | |
| Inversion | Chr5 | 21391219 | 21391532 | LOC_Os05g36090 |
| Inversion | Chr1 | 21420384 | 21420615 | LOC_Os01g38229 |
| Inversion | Chr7 | 21477234 | 21477673 | |
| Inversion | Chr3 | 23019959 | 23020143 | |
| Inversion | Chr11 | 23825850 | 23826010 | |
| Inversion | Chr8 | 25201349 | 25201601 | LOC_Os08g39810 |
| Inversion | Chr5 | 27138634 | 27138831 | |
| Inversion | Chr1 | 30989829 | 30990125 | |
| Inversion | Chr2 | 33497224 | 33497589 | LOC_Os02g54710 |
| Inversion | Chr2 | 35759361 | 35759676 | LOC_Os02g58480 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 4607510 | Chr9 | 7511813 | LOC_Os09g13010 |
| Translocation | Chr9 | 8114346 | Chr6 | 23491898 | |
| Translocation | Chr9 | 8294683 | Chr2 | 16818162 | |
| Translocation | Chr11 | 14753919 | Chr6 | 9352429 | |
| Translocation | Chr10 | 15453844 | Chr1 | 20018271 | LOC_Os10g29720 |
| Translocation | Chr10 | 16050414 | Chr7 | 9781385 | LOC_Os07g16660 |
| Translocation | Chr5 | 16961978 | Chr2 | 2674027 | |
| Translocation | Chr12 | 27080746 | Chr6 | 8332724 | |
| Translocation | Chr6 | 30445662 | Chr1 | 41392692 | LOC_Os06g50300 |
