Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN3902-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN3902-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 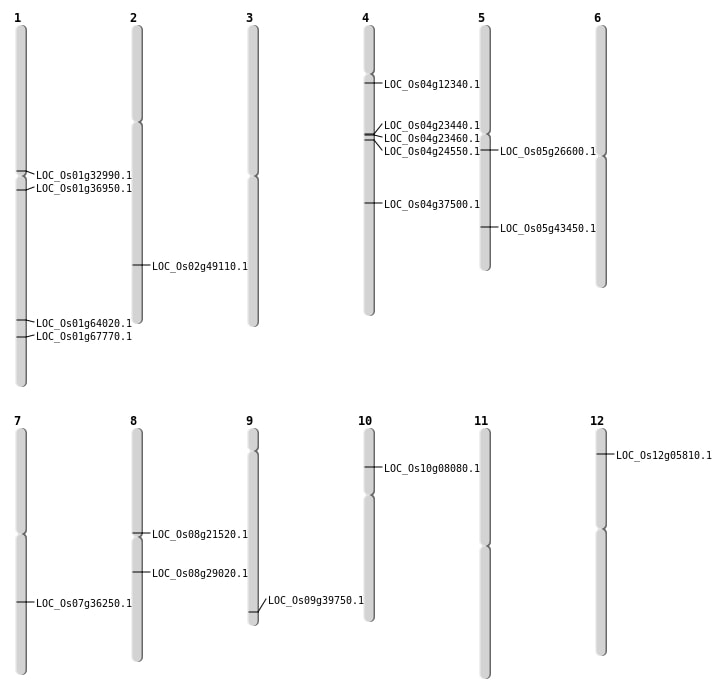
|
| Variant Information |
|---|
Single base substitutions: 49
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 26513128 | C-A | HET | ||
| SBS | Chr1 | 3026804 | T-C | HET | ||
| SBS | Chr1 | 38878551 | T-A | HET | ||
| SBS | Chr1 | 3905569 | T-A | Homo | ||
| SBS | Chr1 | 39389213 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g67770 |
| SBS | Chr10 | 16399902 | C-T | Homo | ||
| SBS | Chr10 | 16399903 | C-T | Homo | ||
| SBS | Chr11 | 10483476 | C-G | HET | ||
| SBS | Chr11 | 15730248 | C-T | HET | ||
| SBS | Chr12 | 2680001 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g05810 |
| SBS | Chr12 | 2680002 | T-A | Homo | ||
| SBS | Chr12 | 4505540 | T-C | Homo | ||
| SBS | Chr12 | 4785637 | T-C | HET | ||
| SBS | Chr12 | 8581098 | A-T | Homo | ||
| SBS | Chr2 | 23890229 | C-T | HET | ||
| SBS | Chr2 | 25068361 | C-G | HET | ||
| SBS | Chr2 | 30033062 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g49110 |
| SBS | Chr2 | 31459376 | G-A | HET | ||
| SBS | Chr2 | 5001701 | T-A | HET | ||
| SBS | Chr2 | 7648414 | A-C | HET | ||
| SBS | Chr3 | 698138 | C-T | HET | ||
| SBS | Chr4 | 14088902 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g24550 |
| SBS | Chr4 | 29884868 | A-G | Homo | ||
| SBS | Chr4 | 6809689 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g12340 |
| SBS | Chr4 | 8058534 | A-G | HET | ||
| SBS | Chr5 | 15456420 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g26600 |
| SBS | Chr5 | 25128559 | C-T | HET | ||
| SBS | Chr5 | 6248957 | G-A | Homo | ||
| SBS | Chr6 | 14270898 | C-T | HET | ||
| SBS | Chr6 | 19965552 | G-A | Homo | ||
| SBS | Chr6 | 21756477 | T-C | Homo | ||
| SBS | Chr7 | 18011209 | G-T | HET | ||
| SBS | Chr7 | 19711841 | T-G | HET | ||
| SBS | Chr7 | 21668739 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g36250 |
| SBS | Chr7 | 25354158 | G-A | HET | ||
| SBS | Chr7 | 29097753 | T-C | HET | ||
| SBS | Chr7 | 3819081 | G-T | HET | ||
| SBS | Chr8 | 12822436 | C-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g21520 |
| SBS | Chr8 | 14990062 | T-A | Homo | ||
| SBS | Chr8 | 17195254 | A-T | HET | ||
| SBS | Chr8 | 17766882 | G-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g29020 |
| SBS | Chr8 | 23056907 | A-T | HET | ||
| SBS | Chr8 | 26903809 | A-T | HET | ||
| SBS | Chr8 | 28167050 | G-A | Homo | ||
| SBS | Chr9 | 17959141 | C-T | HET | ||
| SBS | Chr9 | 18575219 | G-C | HET | ||
| SBS | Chr9 | 22800631 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g39750 |
| SBS | Chr9 | 3750834 | G-T | Homo | ||
| SBS | Chr9 | 9928089 | G-A | Homo |
Deletions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 3873423 | 3873426 | 3 | |
| Deletion | Chr10 | 4377265 | 4377269 | 4 | LOC_Os10g08080 |
| Deletion | Chr12 | 7661168 | 7661172 | 4 | |
| Deletion | Chr8 | 8130054 | 8130056 | 2 | |
| Deletion | Chr1 | 12387764 | 12387769 | 5 | |
| Deletion | Chr8 | 12784043 | 12784048 | 5 | |
| Deletion | Chr4 | 13392001 | 13414000 | 21999 | 2 |
| Deletion | Chr7 | 16435439 | 16435441 | 2 | |
| Deletion | Chr1 | 18116997 | 18117007 | 10 | LOC_Os01g32990 |
| Deletion | Chr1 | 20602131 | 20602133 | 2 | LOC_Os01g36950 |
| Deletion | Chr6 | 21622287 | 21622315 | 28 | |
| Deletion | Chr4 | 22330955 | 22330957 | 2 | LOC_Os04g37500 |
| Deletion | Chr5 | 25261430 | 25261439 | 9 | LOC_Os05g43450 |
| Deletion | Chr2 | 26872551 | 26872554 | 3 | LOC_Os02g44380 |
| Deletion | Chr3 | 27645383 | 27645384 | 1 | |
| Deletion | Chr3 | 28405802 | 28405803 | 1 | |
| Deletion | Chr2 | 32216566 | 32216571 | 5 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 12931417 | 12931418 | 2 | |
| Insertion | Chr1 | 18133783 | 18133783 | 1 | |
| Insertion | Chr4 | 480831 | 480831 | 1 | |
| Insertion | Chr6 | 16046926 | 16046935 | 10 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 24806373 | 25614576 | |
| Inversion | Chr5 | 24806385 | 25614577 | |
| Inversion | Chr1 | 37185027 | 37185370 | LOC_Os01g64020 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 4306960 | Chr6 | 25086676 | |
| Translocation | Chr10 | 17323126 | Chr1 | 23664817 | |
| Translocation | Chr12 | 24332919 | Chr3 | 20216542 | |
| Translocation | Chr4 | 28786005 | Chr3 | 15786019 | |
| Translocation | Chr4 | 28786015 | Chr3 | 15777973 |
