Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN409-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN409-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 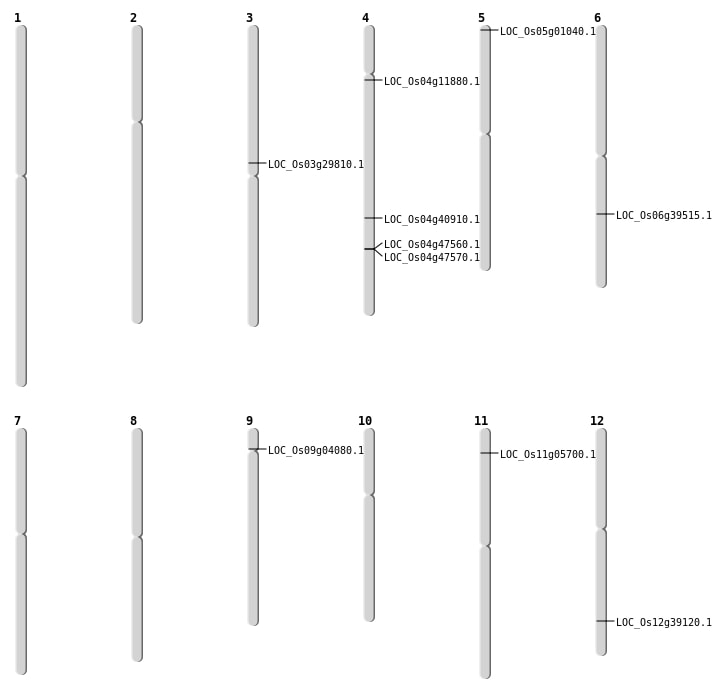
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12215457 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 27873552 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 9782543 | T-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 15936652 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr11 | 17246426 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 2307978 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 2609676 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g05700 |
| SBS | Chr12 | 1371226 | C-T | HET | INTRON | |
| SBS | Chr12 | 24067313 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g39120 |
| SBS | Chr12 | 25214449 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 11502385 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 14182652 | A-C | HET | INTERGENIC | |
| SBS | Chr3 | 14272753 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 16997100 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g29810 |
| SBS | Chr3 | 18817260 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 22061735 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 2628866 | T-G | HET | INTRON | |
| SBS | Chr3 | 35444403 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 21215831 | G-A | HOMO | UTR_3_PRIME | |
| SBS | Chr4 | 24281550 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g40910 |
| SBS | Chr5 | 13252411 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 19926657 | C-G | HET | INTRON | |
| SBS | Chr5 | 2009262 | T-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 21926901 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr5 | 27379408 | T-C | HET | INTRON | |
| SBS | Chr5 | 416525 | T-A | HOMO | INTRON | |
| SBS | Chr5 | 6762351 | T-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 9662277 | C-T | HET | INTRON | |
| SBS | Chr6 | 20692121 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 1049830 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 6783730 | T-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 11592499 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 13816574 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 532431 | C-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 20584643 | C-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 2110542 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g04080 |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 68590 | 68592 | 2 | LOC_Os05g01040 |
| Deletion | Chr11 | 1283926 | 1283941 | 15 | |
| Deletion | Chr12 | 1991929 | 1991934 | 5 | |
| Deletion | Chr12 | 3883649 | 3883653 | 4 | |
| Deletion | Chr7 | 4095483 | 4095517 | 34 | |
| Deletion | Chr6 | 4232409 | 4232414 | 5 | |
| Deletion | Chr3 | 4801149 | 4801163 | 14 | |
| Deletion | Chr4 | 6511832 | 6511850 | 18 | LOC_Os04g11880 |
| Deletion | Chr7 | 8170589 | 8170594 | 5 | |
| Deletion | Chr10 | 13704106 | 13704118 | 12 | |
| Deletion | Chr8 | 13769429 | 13769442 | 13 | |
| Deletion | Chr10 | 13837088 | 13837098 | 10 | |
| Deletion | Chr3 | 14102795 | 14102797 | 2 | |
| Deletion | Chr6 | 14162062 | 14162067 | 5 | |
| Deletion | Chr2 | 14296594 | 14296599 | 5 | |
| Deletion | Chr12 | 16854269 | 16854270 | 1 | |
| Deletion | Chr1 | 21612333 | 21612344 | 11 | |
| Deletion | Chr12 | 24112379 | 24112380 | 1 | |
| Deletion | Chr4 | 28205958 | 28214147 | 8189 | 2 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 26040912 | 26040912 | 1 | |
| Insertion | Chr2 | 22251574 | 22251574 | 1 | |
| Insertion | Chr3 | 7620255 | 7620256 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 6282940 | 7223082 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 23463353 | Chr3 | 29226757 | LOC_Os06g39515 |
| Translocation | Chr11 | 25330902 | Chr7 | 6596893 |
