Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN414-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN414-S Alignment File |
| Seed Availability | No |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 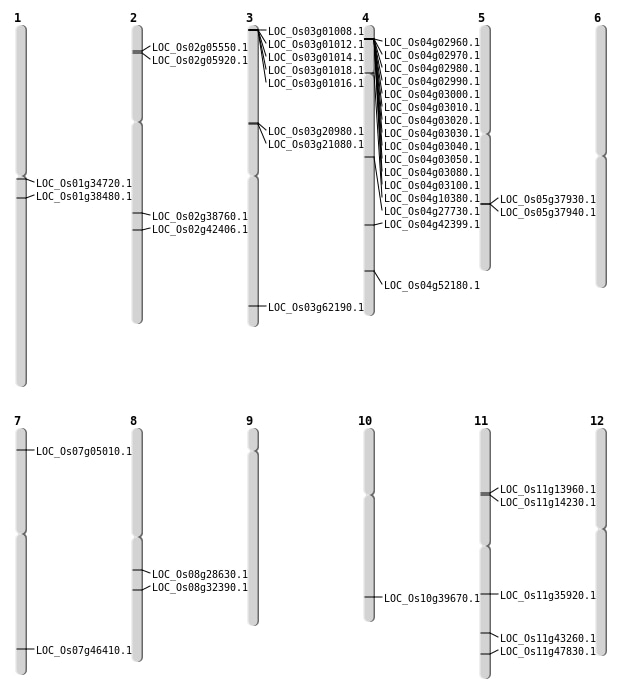
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 27610781 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 16876206 | G-A | HOMO | INTRON | |
| SBS | Chr11 | 10845900 | C-T | HET | INTRON | |
| SBS | Chr11 | 1239364 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 7597021 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 12569877 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 12569880 | C-A | HOMO | UTR_5_PRIME | |
| SBS | Chr2 | 19181468 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 29020687 | T-C | HET | UTR_5_PRIME | |
| SBS | Chr2 | 31919040 | A-T | HOMO | INTRON | |
| SBS | Chr2 | 33687588 | A-C | HET | INTERGENIC | |
| SBS | Chr2 | 34383037 | T-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr2 | 9425374 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 11903888 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g20980 |
| SBS | Chr3 | 200010 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 35225100 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g62190 |
| SBS | Chr4 | 10606396 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 25088817 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g42399 |
| SBS | Chr4 | 26872782 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 30846725 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 34563303 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 5609214 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g10380 |
| SBS | Chr4 | 5609215 | C-A | HET | STOP_GAINED | LOC_Os04g10380 |
| SBS | Chr4 | 8298864 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 9612683 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 11632087 | T-G | HET | INTRON | |
| SBS | Chr5 | 14298692 | A-T | HET | INTRON | |
| SBS | Chr5 | 7275038 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 8718539 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 17673566 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 26169817 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 27695418 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g46410 |
| SBS | Chr8 | 10756048 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 2059577 | A-G | HET | INTERGENIC |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 1001 | 21000 | 20000 | 5 |
| Deletion | Chr4 | 1196001 | 1290000 | 94000 | 12 |
| Deletion | Chr4 | 1496389 | 1496390 | 1 | |
| Deletion | Chr7 | 2210221 | 2210222 | 1 | LOC_Os07g05010 |
| Deletion | Chr2 | 2683493 | 2683506 | 13 | LOC_Os02g05550 |
| Deletion | Chr6 | 2786496 | 2786497 | 1 | |
| Deletion | Chr2 | 2939376 | 2939380 | 4 | LOC_Os02g05920 |
| Deletion | Chr11 | 3277741 | 3277746 | 5 | |
| Deletion | Chr6 | 6416936 | 6416937 | 1 | |
| Deletion | Chr8 | 11287704 | 11287716 | 12 | |
| Deletion | Chr2 | 13085881 | 13085891 | 10 | |
| Deletion | Chr2 | 14071226 | 14071233 | 7 | |
| Deletion | Chr2 | 15201858 | 15201859 | 1 | |
| Deletion | Chr4 | 16388804 | 16388807 | 3 | LOC_Os04g27730 |
| Deletion | Chr2 | 19060828 | 19060834 | 6 | |
| Deletion | Chr8 | 20060886 | 20060887 | 1 | LOC_Os08g32390 |
| Deletion | Chr2 | 20778896 | 20778908 | 12 | |
| Deletion | Chr10 | 21201920 | 21201922 | 2 | LOC_Os10g39670 |
| Deletion | Chr1 | 21626691 | 21626700 | 9 | LOC_Os01g38480 |
| Deletion | Chr5 | 22239480 | 22254715 | 15235 | 2 |
| Deletion | Chr8 | 23230384 | 23230387 | 3 | |
| Deletion | Chr12 | 23829856 | 23829870 | 14 | |
| Deletion | Chr1 | 23989302 | 23989304 | 2 | |
| Deletion | Chr8 | 24351108 | 24351125 | 17 | |
| Deletion | Chr4 | 25364162 | 25364164 | 2 | |
| Deletion | Chr1 | 29172241 | 29172242 | 1 | |
| Deletion | Chr4 | 30999971 | 31001220 | 1249 | LOC_Os04g52180 |
| Deletion | Chr2 | 31932012 | 31932013 | 1 | |
| Deletion | Chr1 | 34007914 | 34007919 | 5 | |
| Deletion | Chr1 | 36444443 | 36444451 | 8 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 18412677 | 18412677 | 1 |
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 7726255 | 7967620 | 2 |
| Inversion | Chr3 | 12006663 | 12554849 | LOC_Os03g21080 |
| Inversion | Chr3 | 12006679 | 12554855 | LOC_Os03g21080 |
| Inversion | Chr11 | 21096575 | 28850603 | 2 |
| Inversion | Chr2 | 31915325 | 32371085 | |
| Inversion | Chr2 | 31915340 | 32371090 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 12049967 | Chr4 | 27823534 | |
| Translocation | Chr10 | 19158654 | Chr2 | 26014056 | |
| Translocation | Chr10 | 19158658 | Chr2 | 25512988 | LOC_Os02g42406 |
| Translocation | Chr5 | 22239490 | Chr3 | 12158520 | LOC_Os05g37930 |
| Translocation | Chr5 | 22254714 | Chr3 | 12158511 | |
| Translocation | Chr11 | 26099974 | Chr8 | 17492313 | 2 |
| Translocation | Chr12 | 26150506 | Chr2 | 23421992 | LOC_Os02g38760 |
| Translocation | Chr11 | 28850604 | Chr1 | 19154520 | 2 |
