Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN429-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN429-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 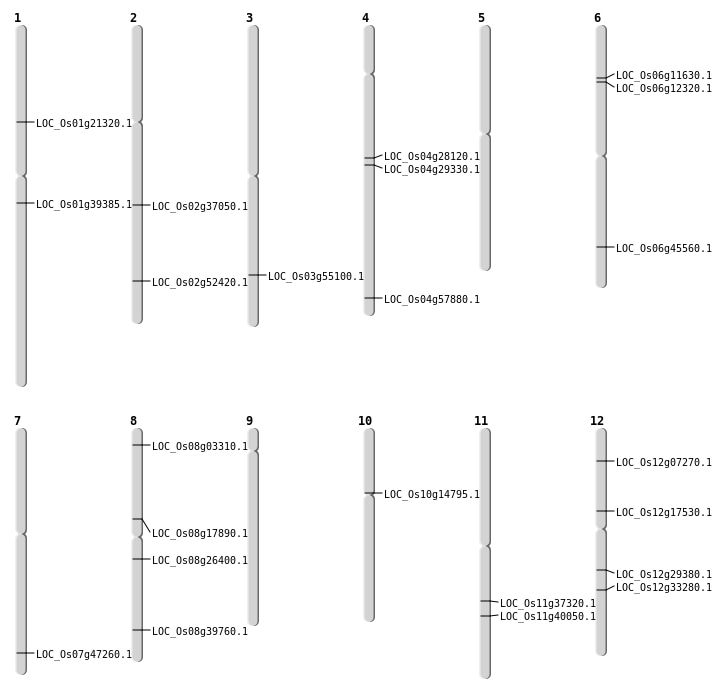
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 2662527 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 3905569 | T-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr1 | 39340831 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 21816941 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 7752556 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g14795 |
| SBS | Chr11 | 18877542 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 21272719 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 22038668 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g37320 |
| SBS | Chr11 | 23881667 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g40050 |
| SBS | Chr12 | 12636112 | T-G | HET | INTRON | |
| SBS | Chr12 | 13078840 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 14283473 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 17457285 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g29380 |
| SBS | Chr12 | 22908954 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 4928108 | C-T | HET | INTRON | |
| SBS | Chr2 | 19744260 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 23630707 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 32081505 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g52420 |
| SBS | Chr3 | 21611196 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 22717 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 30694461 | G-C | HOMO | UTR_3_PRIME | |
| SBS | Chr4 | 17407611 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g29330 |
| SBS | Chr4 | 1926561 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 4381115 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 17177457 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 22099348 | T-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 5295969 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 13847285 | T-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 14857884 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 14259863 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 18011209 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 28247783 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g47260 |
| SBS | Chr7 | 29249124 | A-G | HET | SPLICE_SITE_REGION | |
| SBS | Chr8 | 10982403 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g17890 |
| SBS | Chr8 | 16087853 | G-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 16087856 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 2546531 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 12042302 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 12908568 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 6474717 | C-T | HET | INTERGENIC |
Deletions: 35
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 172088 | 172092 | 4 | |
| Deletion | Chr2 | 708451 | 708495 | 44 | |
| Deletion | Chr8 | 1537042 | 1537046 | 4 | LOC_Os08g03310 |
| Deletion | Chr3 | 3250001 | 3250003 | 2 | |
| Deletion | Chr12 | 3574320 | 3574330 | 10 | LOC_Os12g07270 |
| Deletion | Chr3 | 3909786 | 3909788 | 2 | |
| Deletion | Chr6 | 4793476 | 4793477 | 1 | |
| Deletion | Chr4 | 5348796 | 5348797 | 1 | |
| Deletion | Chr6 | 6161630 | 6161631 | 1 | LOC_Os06g11630 |
| Deletion | Chr6 | 6678861 | 6678865 | 4 | LOC_Os06g12320 |
| Deletion | Chr8 | 8130054 | 8130056 | 2 | |
| Deletion | Chr12 | 10050420 | 10050451 | 31 | LOC_Os12g17530 |
| Deletion | Chr1 | 11909249 | 11909262 | 13 | LOC_Os01g21320 |
| Deletion | Chr11 | 12898408 | 12898412 | 4 | |
| Deletion | Chr12 | 12963530 | 12963539 | 9 | |
| Deletion | Chr3 | 15358422 | 15358432 | 10 | |
| Deletion | Chr4 | 15895711 | 15895797 | 86 | |
| Deletion | Chr8 | 16084549 | 16084550 | 1 | LOC_Os08g26400 |
| Deletion | Chr4 | 16603423 | 16603429 | 6 | LOC_Os04g28120 |
| Deletion | Chr2 | 18880276 | 18880288 | 12 | |
| Deletion | Chr7 | 19995303 | 19995312 | 9 | |
| Deletion | Chr7 | 20360120 | 20360121 | 1 | |
| Deletion | Chr11 | 21242754 | 21242770 | 16 | |
| Deletion | Chr4 | 21526224 | 21526229 | 5 | |
| Deletion | Chr1 | 22195170 | 22195175 | 5 | LOC_Os01g39385 |
| Deletion | Chr2 | 22382683 | 22382686 | 3 | LOC_Os02g37050 |
| Deletion | Chr3 | 22831793 | 22831800 | 7 | |
| Deletion | Chr6 | 24588059 | 24588069 | 10 | |
| Deletion | Chr8 | 25179440 | 25179446 | 6 | LOC_Os08g39760 |
| Deletion | Chr6 | 27589008 | 27589015 | 7 | LOC_Os06g45560 |
| Deletion | Chr3 | 31349668 | 31349681 | 13 | LOC_Os03g55100 |
| Deletion | Chr2 | 32081490 | 32081496 | 6 | LOC_Os02g52420 |
| Deletion | Chr2 | 32216566 | 32216571 | 5 | |
| Deletion | Chr4 | 34490206 | 34490212 | 6 | LOC_Os04g57880 |
| Deletion | Chr1 | 39135892 | 39135901 | 9 |
Insertions: 5
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 24719628 | 24939848 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 10453751 | Chr2 | 12969273 | |
| Translocation | Chr11 | 18280684 | Chr3 | 29622228 | |
| Translocation | Chr12 | 18702617 | Chr11 | 23936814 | |
| Translocation | Chr12 | 20138349 | Chr5 | 13277467 | LOC_Os12g33280 |
