Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4353-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4353-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 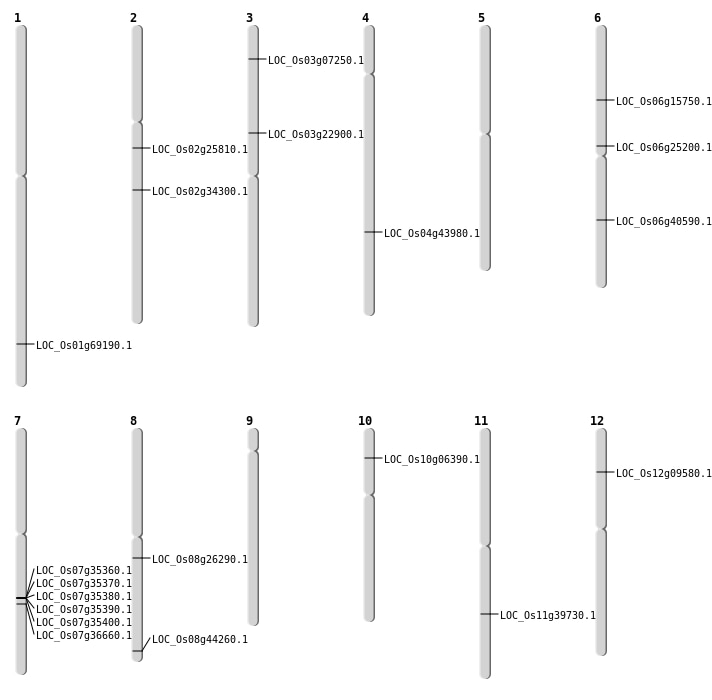
|
| Variant Information |
|---|
Single base substitutions: 71
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17197523 | G-C | HET | UTR_3_PRIME | |
| SBS | Chr1 | 35190692 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 35555868 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 38342486 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 39886717 | T-C | HET | SPLICE_SITE_REGION | |
| SBS | Chr1 | 42028501 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 10740889 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 3284272 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g06390 |
| SBS | Chr10 | 659077 | T-G | HET | INTERGENIC | |
| SBS | Chr11 | 7896117 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 8776407 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 20434190 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 23592551 | G-A | HET | INTRON | |
| SBS | Chr12 | 5039362 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g09580 |
| SBS | Chr2 | 15125323 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g25810 |
| SBS | Chr2 | 20526845 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g34300 |
| SBS | Chr2 | 28691744 | T-C | HET | INTRON | |
| SBS | Chr2 | 31912908 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 31912909 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 10113212 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 13239227 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g22900 |
| SBS | Chr3 | 1708817 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 22001597 | G-T | HET | UTR_3_PRIME | |
| SBS | Chr3 | 26734498 | C-A | HET | INTRON | |
| SBS | Chr3 | 29362221 | G-A | HET | SPLICE_SITE_REGION | |
| SBS | Chr4 | 12142165 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 12919458 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 21173096 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 2361885 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 23773601 | A-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 25685472 | C-T | HET | INTRON | |
| SBS | Chr4 | 26070235 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g43980 |
| SBS | Chr4 | 30611881 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 33515887 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 3363888 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 33838209 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 5702070 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 7279312 | G-A | HOMO | INTRON | |
| SBS | Chr4 | 8032388 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 15415732 | A-T | HOMO | INTRON | |
| SBS | Chr5 | 15655138 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 29342508 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 14741277 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g25200 |
| SBS | Chr6 | 15626606 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 16369018 | A-C | HET | INTERGENIC | |
| SBS | Chr6 | 5274614 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 8938825 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g15750 |
| SBS | Chr7 | 10563398 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 15156479 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 18620776 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 21940266 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g36660 |
| SBS | Chr7 | 7825997 | A-G | HET | INTRON | |
| SBS | Chr7 | 8145874 | G-A | HET | UTR_5_PRIME | |
| SBS | Chr8 | 16027131 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g26290 |
| SBS | Chr8 | 16638133 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 17718592 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 17719299 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 17980502 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 20550625 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 25162438 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 25732827 | A-T | HET | INTRON | |
| SBS | Chr8 | 27752008 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 27865223 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g44260 |
| SBS | Chr8 | 5219977 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 1952426 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 20101828 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 21117381 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 22420447 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 4105020 | T-C | HET | INTRON | |
| SBS | Chr9 | 4826850 | G-A | HET | INTRON | |
| SBS | Chr9 | 4827813 | T-C | HET | INTRON |
Deletions: 35
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 1755248 | 1755268 | 20 | |
| Deletion | Chr5 | 2963867 | 2963885 | 18 | |
| Deletion | Chr3 | 3702827 | 3702828 | 1 | LOC_Os03g07250 |
| Deletion | Chr5 | 3833839 | 3833840 | 1 | |
| Deletion | Chr1 | 4146450 | 4146454 | 4 | |
| Deletion | Chr11 | 5098744 | 5098817 | 73 | |
| Deletion | Chr12 | 5351003 | 5351005 | 2 | |
| Deletion | Chr1 | 6428083 | 6428098 | 15 | |
| Deletion | Chr9 | 7335141 | 7335239 | 98 | |
| Deletion | Chr9 | 7946021 | 7946031 | 10 | |
| Deletion | Chr7 | 9097579 | 9097586 | 7 | |
| Deletion | Chr5 | 9750747 | 9750748 | 1 | |
| Deletion | Chr12 | 10241841 | 10241842 | 1 | |
| Deletion | Chr5 | 10658557 | 10658560 | 3 | |
| Deletion | Chr3 | 10795276 | 10795277 | 1 | |
| Deletion | Chr11 | 11336425 | 11336436 | 11 | |
| Deletion | Chr7 | 11623766 | 11623777 | 11 | |
| Deletion | Chr12 | 12280278 | 12280287 | 9 | |
| Deletion | Chr10 | 12797538 | 12797600 | 62 | |
| Deletion | Chr10 | 13384828 | 13384829 | 1 | |
| Deletion | Chr1 | 13770708 | 13770763 | 55 | |
| Deletion | Chr1 | 16323546 | 16323547 | 1 | |
| Deletion | Chr10 | 17526473 | 17526474 | 1 | |
| Deletion | Chr5 | 18371837 | 18371838 | 1 | |
| Deletion | Chr12 | 18402478 | 18402479 | 1 | |
| Deletion | Chr2 | 19419067 | 19419144 | 77 | |
| Deletion | Chr7 | 21153001 | 21174000 | 21000 | 5 |
| Deletion | Chr11 | 21457696 | 21457701 | 5 | |
| Deletion | Chr11 | 21462437 | 21462449 | 12 | |
| Deletion | Chr9 | 21618822 | 21618824 | 2 | |
| Deletion | Chr11 | 23677171 | 23677172 | 1 | LOC_Os11g39730 |
| Deletion | Chr12 | 24016480 | 24016482 | 2 | |
| Deletion | Chr4 | 25970961 | 25970971 | 10 | |
| Deletion | Chr1 | 29476577 | 29476580 | 3 | |
| Deletion | Chr1 | 40211374 | 40211375 | 1 | LOC_Os01g69190 |
Insertions: 12
Inversions: 5
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 17927239 | Chr1 | 16605896 | |
| Translocation | Chr6 | 24203346 | Chr4 | 18789587 | |
| Translocation | Chr6 | 24203577 | Chr4 | 18789577 | LOC_Os06g40590 |
