Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4395-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4395-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 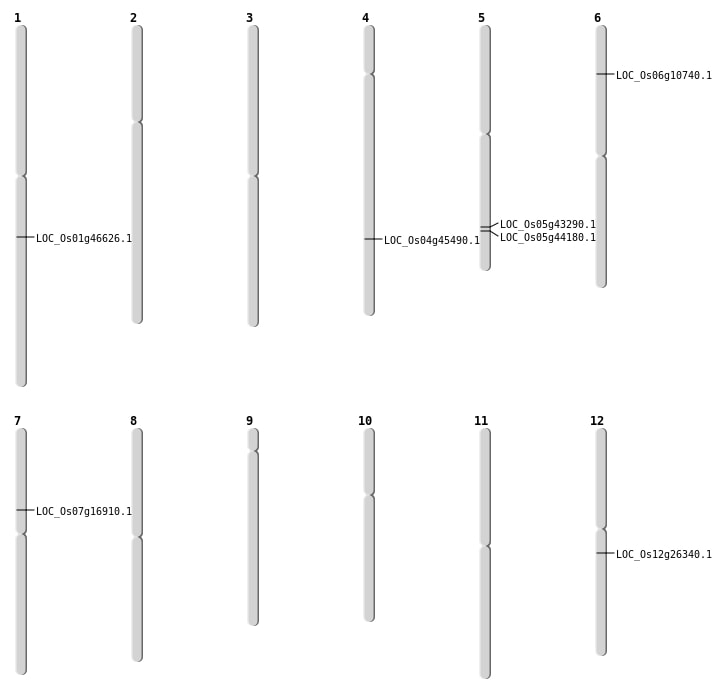
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10662999 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 15637406 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 21944288 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr1 | 25427339 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 26548216 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g46626 |
| SBS | Chr1 | 33804098 | A-G | HET | INTRON | |
| SBS | Chr10 | 11791873 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 16882081 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 23648900 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 3897504 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 15389665 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 15389667 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g26340 |
| SBS | Chr12 | 21433247 | C-G | HOMO | INTRON | |
| SBS | Chr2 | 10904835 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 14962313 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr2 | 24743571 | G-A | HET | INTRON | |
| SBS | Chr2 | 6168335 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 22821183 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 33585325 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 13696780 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 14402308 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 18147836 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 11520923 | C-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 1013860 | C-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 12662024 | C-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 20142081 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 21655537 | T-C | HOMO | INTRON | |
| SBS | Chr6 | 6327940 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 7023334 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 9949458 | G-A | HET | STOP_GAINED | LOC_Os07g16910 |
| SBS | Chr8 | 10523259 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 6336932 | T-C | HET | INTRON |
Deletions: 37
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 714918 | 714919 | 1 | |
| Deletion | Chr9 | 2449714 | 2449715 | 1 | |
| Deletion | Chr8 | 3004145 | 3004146 | 1 | |
| Deletion | Chr4 | 3503148 | 3503149 | 1 | |
| Deletion | Chr5 | 3649006 | 3649012 | 6 | |
| Deletion | Chr5 | 3661946 | 3661962 | 16 | |
| Deletion | Chr7 | 4174071 | 4174072 | 1 | |
| Deletion | Chr11 | 4923974 | 4923976 | 2 | |
| Deletion | Chr10 | 5081996 | 5081997 | 1 | |
| Deletion | Chr6 | 5610574 | 5610575 | 1 | LOC_Os06g10740 |
| Deletion | Chr8 | 6443606 | 6443634 | 28 | |
| Deletion | Chr12 | 7385920 | 7385922 | 2 | |
| Deletion | Chr12 | 7851717 | 7851721 | 4 | |
| Deletion | Chr12 | 9106956 | 9106958 | 2 | |
| Deletion | Chr12 | 9977351 | 9977358 | 7 | |
| Deletion | Chr7 | 10134930 | 10134932 | 2 | |
| Deletion | Chr2 | 11186660 | 11186662 | 2 | |
| Deletion | Chr6 | 12006873 | 12006889 | 16 | |
| Deletion | Chr5 | 14281081 | 14281084 | 3 | |
| Deletion | Chr6 | 14715432 | 14715433 | 1 | |
| Deletion | Chr3 | 14781870 | 14781887 | 17 | |
| Deletion | Chr2 | 15713348 | 15713351 | 3 | |
| Deletion | Chr8 | 15888081 | 15888082 | 1 | |
| Deletion | Chr7 | 17464001 | 17464002 | 1 | |
| Deletion | Chr3 | 17515041 | 17515042 | 1 | |
| Deletion | Chr12 | 18996140 | 18996141 | 1 | |
| Deletion | Chr12 | 19491864 | 19491867 | 3 | |
| Deletion | Chr10 | 20270570 | 20270579 | 9 | |
| Deletion | Chr11 | 20495143 | 20495144 | 1 | |
| Deletion | Chr8 | 21971791 | 21971792 | 1 | |
| Deletion | Chr2 | 23510882 | 23510883 | 1 | |
| Deletion | Chr6 | 24458069 | 24458074 | 5 | |
| Deletion | Chr12 | 24587178 | 24587193 | 15 | |
| Deletion | Chr5 | 25667449 | 25667459 | 10 | LOC_Os05g44180 |
| Deletion | Chr7 | 27764606 | 27764674 | 68 | |
| Deletion | Chr5 | 29795895 | 29795899 | 4 | |
| Deletion | Chr6 | 29797737 | 29797738 | 1 |
Insertions: 10
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 1314466 | 1314481 | 16 | |
| Insertion | Chr1 | 396366 | 396367 | 2 | |
| Insertion | Chr11 | 4801321 | 4801321 | 1 | |
| Insertion | Chr11 | 9981486 | 9981486 | 1 | |
| Insertion | Chr3 | 19577286 | 19577286 | 1 | |
| Insertion | Chr3 | 28016786 | 28016787 | 2 | |
| Insertion | Chr4 | 26920556 | 26920636 | 81 | LOC_Os04g45490 |
| Insertion | Chr5 | 500547 | 500548 | 2 | |
| Insertion | Chr6 | 8170442 | 8170456 | 15 | |
| Insertion | Chr9 | 20303192 | 20303192 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 25187034 | 25352911 | LOC_Os05g43290 |
| Inversion | Chr5 | 25187044 | 25352921 |
No Translocation
