Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4405-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4405-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 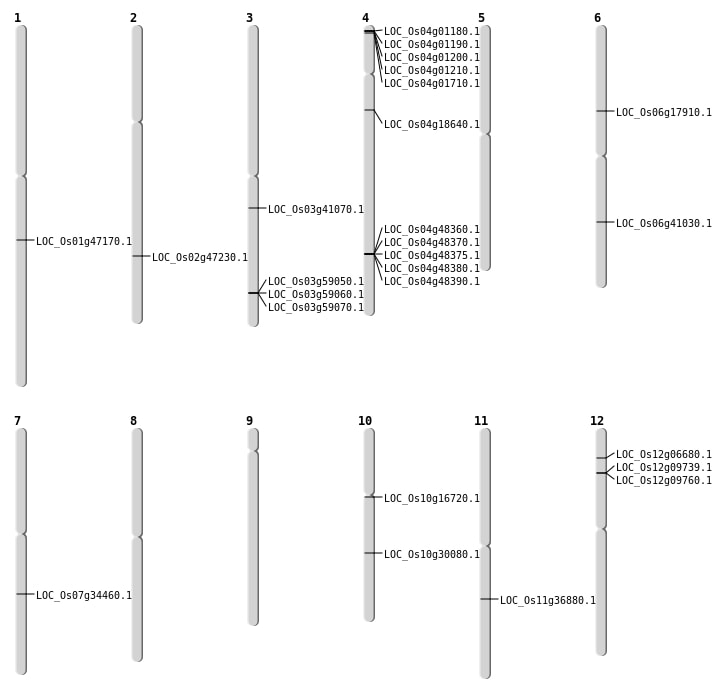
|
| Variant Information |
|---|
Single base substitutions: 44
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 22045649 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 26963525 | G-A | HET | STOP_GAINED | LOC_Os01g47170 |
| SBS | Chr1 | 31793625 | C-G | HET | INTERGENIC | |
| SBS | Chr1 | 32097511 | T-A | HET | INTRON | |
| SBS | Chr1 | 35556551 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 38121089 | A-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 40684332 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 5213622 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 17684781 | G-A | HET | INTRON | |
| SBS | Chr10 | 8331442 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g16720 |
| SBS | Chr11 | 21758936 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g36880 |
| SBS | Chr11 | 9295729 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 9295730 | A-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 763826 | A-G | HET | UTR_5_PRIME | |
| SBS | Chr2 | 10835296 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 10835298 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 14153795 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 24968914 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 14371092 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 22826219 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g41070 |
| SBS | Chr3 | 23997617 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 24505830 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 36251740 | T-C | HET | INTRON | |
| SBS | Chr3 | 7677076 | T-C | HOMO | UTR_5_PRIME | |
| SBS | Chr4 | 10305428 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 10305429 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g18640 |
| SBS | Chr4 | 19291667 | C-T | HET | INTRON | |
| SBS | Chr4 | 19748827 | G-A | HET | INTRON | |
| SBS | Chr4 | 31515190 | C-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 34449109 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 15370221 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 18486165 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 21536851 | C-T | HET | INTRON | |
| SBS | Chr5 | 9911059 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 10398621 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g17910 |
| SBS | Chr6 | 16883964 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 17584727 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 24063811 | G-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 25442140 | C-T | HOMO | INTRON | |
| SBS | Chr7 | 25442141 | C-T | HOMO | INTRON | |
| SBS | Chr8 | 13850808 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 28419710 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 4258507 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 5645227 | C-G | HET | SYNONYMOUS_CODING |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 133001 | 162000 | 29000 | 4 |
| Deletion | Chr4 | 472483 | 472487 | 4 | LOC_Os04g01710 |
| Deletion | Chr5 | 2349886 | 2349891 | 5 | |
| Deletion | Chr9 | 2373010 | 2373011 | 1 | |
| Deletion | Chr7 | 3046169 | 3046180 | 11 | |
| Deletion | Chr12 | 3253054 | 3253064 | 10 | LOC_Os12g06680 |
| Deletion | Chr10 | 4560409 | 4560410 | 1 | |
| Deletion | Chr4 | 4775112 | 4775123 | 11 | |
| Deletion | Chr12 | 5143001 | 5171000 | 28000 | 2 |
| Deletion | Chr5 | 5936905 | 5936906 | 1 | |
| Deletion | Chr1 | 6454154 | 6454157 | 3 | |
| Deletion | Chr4 | 8135675 | 8135678 | 3 | |
| Deletion | Chr9 | 9848752 | 9848760 | 8 | |
| Deletion | Chr5 | 10597715 | 10597717 | 2 | |
| Deletion | Chr10 | 15624044 | 15624053 | 9 | LOC_Os10g30080 |
| Deletion | Chr7 | 17804335 | 17804336 | 1 | |
| Deletion | Chr11 | 18672532 | 18672535 | 3 | |
| Deletion | Chr1 | 20401969 | 20401985 | 16 | |
| Deletion | Chr8 | 22422706 | 22422707 | 1 | |
| Deletion | Chr1 | 23620500 | 23620567 | 67 | |
| Deletion | Chr3 | 23876614 | 23876623 | 9 | |
| Deletion | Chr7 | 25631696 | 25631699 | 3 | |
| Deletion | Chr6 | 25663260 | 25663265 | 5 | |
| Deletion | Chr3 | 27118963 | 27118965 | 2 | |
| Deletion | Chr4 | 28826001 | 28869000 | 43000 | 5 |
| Deletion | Chr2 | 31922863 | 31922864 | 1 | |
| Deletion | Chr4 | 32588588 | 32588597 | 9 | |
| Deletion | Chr3 | 33538764 | 33538770 | 6 | LOC_Os03g58910 |
| Deletion | Chr3 | 33615001 | 33631000 | 16000 | 3 |
Insertions: 7
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 22659914 | 22659915 | 2 | |
| Insertion | Chr11 | 159877 | 159913 | 37 | |
| Insertion | Chr2 | 5636488 | 5636488 | 1 | |
| Insertion | Chr7 | 20663169 | 20663169 | 1 | LOC_Os07g34460 |
| Insertion | Chr7 | 21633553 | 21633553 | 1 | |
| Insertion | Chr7 | 9981329 | 9981342 | 14 | |
| Insertion | Chr9 | 18313692 | 18313703 | 12 |
Inversions: 11
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 3136781 | 3136880 | |
| Inversion | Chr12 | 6994632 | 7015054 | |
| Inversion | Chr6 | 14787878 | 14788038 | |
| Inversion | Chr11 | 14788412 | 14788652 | |
| Inversion | Chr5 | 23222477 | 23222586 | |
| Inversion | Chr6 | 24192791 | 24503176 | LOC_Os06g41030 |
| Inversion | Chr6 | 24192803 | 24503185 | LOC_Os06g41030 |
| Inversion | Chr11 | 27711872 | 27712104 | |
| Inversion | Chr5 | 28196703 | 28196837 | |
| Inversion | Chr2 | 30677010 | 31543800 | |
| Inversion | Chr2 | 30686467 | 31543822 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 166703 | Chr2 | 15134549 | |
| Translocation | Chr4 | 28827023 | Chr2 | 15134547 | LOC_Os02g47230 |
| Translocation | Chr4 | 28868945 | Chr2 | 15134159 | LOC_Os04g48390 |
