Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4449-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4449-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 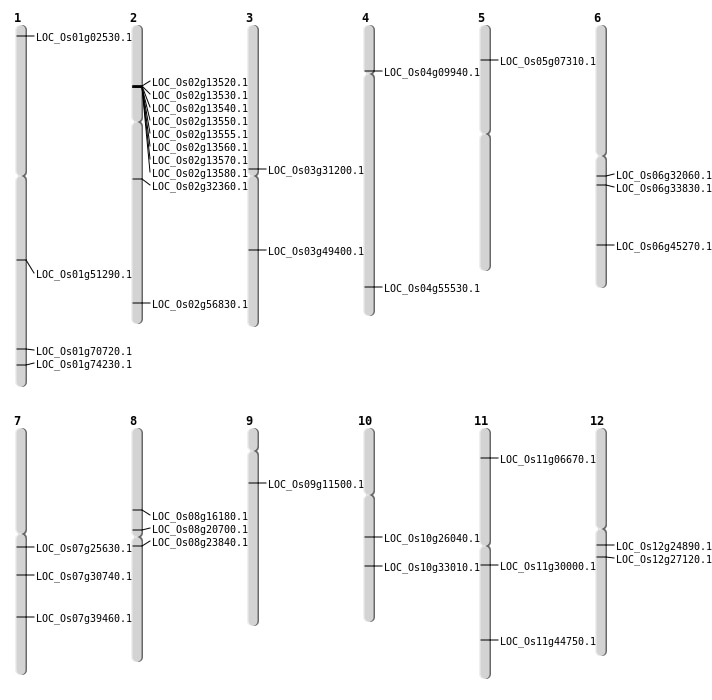
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15701489 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 827287 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g02530 |
| SBS | Chr11 | 13147258 | G-A | HET | INTRON | |
| SBS | Chr11 | 14083774 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 17440195 | G-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g30000 |
| SBS | Chr11 | 22104033 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 27088360 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g44750 |
| SBS | Chr12 | 22449055 | G-A | HET | INTRON | |
| SBS | Chr2 | 19112550 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g32360 |
| SBS | Chr2 | 20228279 | G-A | HOMO | INTRON | |
| SBS | Chr3 | 10134183 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 18235260 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 18235261 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 28121513 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g49400 |
| SBS | Chr3 | 28121514 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 4467224 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 10301809 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 23752745 | A-T | HOMO | UTR_5_PRIME | |
| SBS | Chr4 | 23752746 | G-T | HOMO | UTR_5_PRIME | |
| SBS | Chr4 | 26845606 | A-G | HET | INTRON | |
| SBS | Chr5 | 22176739 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 23979148 | A-G | HET | INTRON | |
| SBS | Chr6 | 12945310 | C-A | HET | INTRON | |
| SBS | Chr6 | 19684845 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g33830 |
| SBS | Chr6 | 20793854 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 23227396 | T-A | HET | INTRON | |
| SBS | Chr6 | 27504575 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 29211191 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 20964053 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 793143 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 10629545 | T-G | HOMO | INTRON | |
| SBS | Chr8 | 12441510 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g20700 |
| SBS | Chr9 | 12671266 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 13247897 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 22841025 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 8728744 | A-T | HOMO | INTRON |
Deletions: 101
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 63054 | 63056 | 2 | |
| Deletion | Chr6 | 110818 | 110819 | 1 | |
| Deletion | Chr11 | 330432 | 330433 | 1 | |
| Deletion | Chr6 | 450384 | 450385 | 1 | |
| Deletion | Chr3 | 899961 | 899962 | 1 | |
| Deletion | Chr2 | 934414 | 934418 | 4 | |
| Deletion | Chr7 | 1004006 | 1004007 | 1 | |
| Deletion | Chr2 | 1351432 | 1351439 | 7 | |
| Deletion | Chr2 | 1358874 | 1358890 | 16 | |
| Deletion | Chr9 | 3390118 | 3390119 | 1 | |
| Deletion | Chr10 | 3505707 | 3505708 | 1 | |
| Deletion | Chr12 | 3518564 | 3518565 | 1 | |
| Deletion | Chr10 | 3555803 | 3555804 | 1 | |
| Deletion | Chr11 | 3984043 | 3984044 | 1 | |
| Deletion | Chr9 | 4294966 | 4294968 | 2 | |
| Deletion | Chr10 | 4567032 | 4567033 | 1 | |
| Deletion | Chr10 | 5019579 | 5019585 | 6 | |
| Deletion | Chr1 | 5211718 | 5211719 | 1 | |
| Deletion | Chr4 | 5345561 | 5345573 | 12 | LOC_Os04g09940 |
| Deletion | Chr6 | 5876529 | 5876530 | 1 | |
| Deletion | Chr4 | 6182618 | 6182619 | 1 | |
| Deletion | Chr8 | 6369429 | 6369430 | 1 | |
| Deletion | Chr11 | 6396226 | 6396264 | 38 | |
| Deletion | Chr3 | 6414598 | 6414599 | 1 | |
| Deletion | Chr9 | 6416295 | 6416296 | 1 | LOC_Os09g11500 |
| Deletion | Chr3 | 6477461 | 6477463 | 2 | |
| Deletion | Chr8 | 6927215 | 6927216 | 1 | |
| Deletion | Chr4 | 7116361 | 7116363 | 2 | |
| Deletion | Chr2 | 7239001 | 7284000 | 45000 | 8 |
| Deletion | Chr12 | 7456734 | 7456735 | 1 | |
| Deletion | Chr11 | 7647785 | 7647786 | 1 | |
| Deletion | Chr5 | 7668599 | 7668600 | 1 | |
| Deletion | Chr1 | 7903367 | 7903368 | 1 | |
| Deletion | Chr10 | 8114330 | 8114352 | 22 | |
| Deletion | Chr2 | 8220608 | 8220691 | 83 | |
| Deletion | Chr8 | 8362667 | 8362668 | 1 | |
| Deletion | Chr5 | 9848209 | 9848211 | 2 | |
| Deletion | Chr8 | 9866852 | 9866853 | 1 | LOC_Os08g16180 |
| Deletion | Chr8 | 9929270 | 9929271 | 1 | |
| Deletion | Chr4 | 10026350 | 10026351 | 1 | |
| Deletion | Chr10 | 10339461 | 10339462 | 1 | |
| Deletion | Chr4 | 10868421 | 10868440 | 19 | |
| Deletion | Chr12 | 11004110 | 11004111 | 1 | |
| Deletion | Chr6 | 11687914 | 11687915 | 1 | |
| Deletion | Chr5 | 11750352 | 11750353 | 1 | |
| Deletion | Chr7 | 11842300 | 11842357 | 57 | |
| Deletion | Chr1 | 12538446 | 12538469 | 23 | |
| Deletion | Chr4 | 12894619 | 12894620 | 1 | |
| Deletion | Chr4 | 13117972 | 13117975 | 3 | |
| Deletion | Chr10 | 13490234 | 13490235 | 1 | LOC_Os10g26040 |
| Deletion | Chr8 | 13594638 | 13594639 | 1 | |
| Deletion | Chr8 | 13636443 | 13636490 | 47 | |
| Deletion | Chr4 | 14166707 | 14166708 | 1 | |
| Deletion | Chr9 | 14230796 | 14230797 | 1 | |
| Deletion | Chr3 | 14256100 | 14256102 | 2 | |
| Deletion | Chr5 | 14559802 | 14559803 | 1 | |
| Deletion | Chr7 | 14696082 | 14696083 | 1 | LOC_Os07g25630 |
| Deletion | Chr8 | 14705882 | 14705885 | 3 | |
| Deletion | Chr6 | 14976864 | 14976874 | 10 | |
| Deletion | Chr8 | 15235841 | 15235842 | 1 | |
| Deletion | Chr10 | 15254659 | 15254660 | 1 | |
| Deletion | Chr8 | 15303552 | 15303562 | 10 | |
| Deletion | Chr8 | 15427187 | 15427188 | 1 | |
| Deletion | Chr12 | 15430960 | 15430962 | 2 | |
| Deletion | Chr2 | 15465818 | 15465822 | 4 | |
| Deletion | Chr4 | 15883567 | 15883568 | 1 | |
| Deletion | Chr12 | 15917231 | 15917233 | 2 | LOC_Os12g27120 |
| Deletion | Chr6 | 16020950 | 16020952 | 2 | |
| Deletion | Chr9 | 16688905 | 16688906 | 1 | |
| Deletion | Chr2 | 17147683 | 17147684 | 1 | |
| Deletion | Chr10 | 17285826 | 17285849 | 23 | LOC_Os10g33010 |
| Deletion | Chr3 | 17767447 | 17767450 | 3 | LOC_Os03g31200 |
| Deletion | Chr3 | 17945406 | 17945407 | 1 | |
| Deletion | Chr7 | 19324814 | 19324815 | 1 | |
| Deletion | Chr12 | 19523833 | 19523839 | 6 | |
| Deletion | Chr8 | 19525714 | 19525715 | 1 | |
| Deletion | Chr5 | 19536900 | 19536901 | 1 | |
| Deletion | Chr1 | 19646812 | 19646816 | 4 | |
| Deletion | Chr11 | 21078793 | 21078794 | 1 | |
| Deletion | Chr7 | 21271320 | 21271322 | 2 | |
| Deletion | Chr2 | 21353744 | 21353745 | 1 | |
| Deletion | Chr6 | 21368303 | 21368304 | 1 | |
| Deletion | Chr6 | 21584939 | 21584940 | 1 | |
| Deletion | Chr6 | 21926778 | 21926779 | 1 | |
| Deletion | Chr4 | 22767490 | 22767491 | 1 | |
| Deletion | Chr2 | 23212273 | 23212274 | 1 | |
| Deletion | Chr11 | 23654403 | 23654405 | 2 | |
| Deletion | Chr8 | 23816574 | 23816575 | 1 | |
| Deletion | Chr5 | 24045519 | 24045520 | 1 | |
| Deletion | Chr6 | 24056668 | 24056669 | 1 | |
| Deletion | Chr6 | 24163428 | 24163434 | 6 | |
| Deletion | Chr12 | 25668526 | 25668533 | 7 | |
| Deletion | Chr1 | 26334509 | 26334527 | 18 | |
| Deletion | Chr6 | 28263849 | 28263850 | 1 | |
| Deletion | Chr1 | 29486902 | 29486905 | 3 | LOC_Os01g51290 |
| Deletion | Chr2 | 31336745 | 31336780 | 35 | |
| Deletion | Chr1 | 32403175 | 32403179 | 4 | |
| Deletion | Chr2 | 34828664 | 34828695 | 31 | LOC_Os02g56830 |
| Deletion | Chr1 | 37614432 | 37614433 | 1 | |
| Deletion | Chr1 | 40603143 | 40603144 | 1 | |
| Deletion | Chr1 | 41659181 | 41659182 | 1 |
Insertions: 90
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 10087630 | 10087630 | 1 | |
| Insertion | Chr1 | 18776225 | 18776230 | 6 | |
| Insertion | Chr1 | 2148084 | 2148084 | 1 | |
| Insertion | Chr1 | 30351652 | 30351652 | 1 | |
| Insertion | Chr1 | 32157225 | 32157225 | 1 | |
| Insertion | Chr1 | 3769371 | 3769371 | 1 | |
| Insertion | Chr1 | 38858773 | 38858773 | 1 | |
| Insertion | Chr1 | 40939340 | 40939340 | 1 | LOC_Os01g70720 |
| Insertion | Chr1 | 41786805 | 41786805 | 1 | |
| Insertion | Chr1 | 43000711 | 43000711 | 1 | LOC_Os01g74230 |
| Insertion | Chr10 | 12168206 | 12168206 | 1 | |
| Insertion | Chr10 | 12477069 | 12477069 | 1 | |
| Insertion | Chr10 | 16822796 | 16822796 | 1 | |
| Insertion | Chr10 | 1770 | 1770 | 1 | |
| Insertion | Chr10 | 20652040 | 20652040 | 1 | |
| Insertion | Chr10 | 342815 | 342815 | 1 | |
| Insertion | Chr10 | 8388381 | 8388381 | 1 | |
| Insertion | Chr11 | 21415784 | 21415784 | 1 | |
| Insertion | Chr11 | 25683123 | 25683123 | 1 | |
| Insertion | Chr11 | 2662104 | 2662105 | 2 | |
| Insertion | Chr11 | 27044991 | 27044991 | 1 | |
| Insertion | Chr11 | 28105288 | 28105291 | 4 | |
| Insertion | Chr12 | 11007901 | 11007902 | 2 | |
| Insertion | Chr12 | 12059218 | 12059218 | 1 | |
| Insertion | Chr12 | 12663077 | 12663077 | 1 | |
| Insertion | Chr12 | 14279212 | 14279212 | 1 | LOC_Os12g24890 |
| Insertion | Chr12 | 14321926 | 14321927 | 2 | |
| Insertion | Chr12 | 14877261 | 14877261 | 1 | |
| Insertion | Chr2 | 14840393 | 14840393 | 1 | |
| Insertion | Chr2 | 17825854 | 17825854 | 1 | |
| Insertion | Chr2 | 32483158 | 32483158 | 1 | |
| Insertion | Chr2 | 379494 | 379494 | 1 | |
| Insertion | Chr2 | 919991 | 919991 | 1 | |
| Insertion | Chr2 | 9354444 | 9354444 | 1 | |
| Insertion | Chr3 | 13469533 | 13469534 | 2 | |
| Insertion | Chr3 | 18742233 | 18742234 | 2 | |
| Insertion | Chr3 | 19130870 | 19130870 | 1 | |
| Insertion | Chr3 | 288499 | 288499 | 1 | |
| Insertion | Chr3 | 31582364 | 31582365 | 2 | |
| Insertion | Chr3 | 3783478 | 3783479 | 2 | |
| Insertion | Chr3 | 4273598 | 4273598 | 1 | |
| Insertion | Chr3 | 6617328 | 6617387 | 60 | |
| Insertion | Chr3 | 9389501 | 9389501 | 1 | |
| Insertion | Chr4 | 11721733 | 11721733 | 1 | |
| Insertion | Chr4 | 13757532 | 13757532 | 1 | |
| Insertion | Chr4 | 19386259 | 19386259 | 1 | |
| Insertion | Chr4 | 19532256 | 19532258 | 3 | |
| Insertion | Chr4 | 20095630 | 20095630 | 1 | |
| Insertion | Chr4 | 22986098 | 22986098 | 1 | |
| Insertion | Chr4 | 24227899 | 24227899 | 1 | |
| Insertion | Chr4 | 2586849 | 2586849 | 1 | |
| Insertion | Chr4 | 2631656 | 2631656 | 1 | |
| Insertion | Chr4 | 30486729 | 30486729 | 1 | |
| Insertion | Chr4 | 31202371 | 31202371 | 1 | |
| Insertion | Chr4 | 33035188 | 33035188 | 1 | LOC_Os04g55530 |
| Insertion | Chr4 | 8954481 | 8954481 | 1 | |
| Insertion | Chr4 | 9227864 | 9227864 | 1 | |
| Insertion | Chr5 | 20358272 | 20358273 | 2 | |
| Insertion | Chr5 | 26351526 | 26351529 | 4 | |
| Insertion | Chr5 | 3884937 | 3884937 | 1 | LOC_Os05g07310 |
| Insertion | Chr5 | 8030655 | 8030655 | 1 | |
| Insertion | Chr5 | 9730801 | 9730847 | 47 | |
| Insertion | Chr6 | 17169333 | 17169333 | 1 | |
| Insertion | Chr6 | 18648236 | 18648236 | 1 | LOC_Os06g32060 |
| Insertion | Chr6 | 2111442 | 2111443 | 2 | |
| Insertion | Chr6 | 24613610 | 24613613 | 4 | |
| Insertion | Chr6 | 27357948 | 27357954 | 7 | LOC_Os06g45270 |
| Insertion | Chr6 | 27380786 | 27380786 | 1 | |
| Insertion | Chr6 | 4802 | 4802 | 1 | |
| Insertion | Chr6 | 5789500 | 5789501 | 2 | |
| Insertion | Chr7 | 18195682 | 18195682 | 1 | LOC_Os07g30740 |
| Insertion | Chr7 | 26147412 | 26147416 | 5 | |
| Insertion | Chr7 | 3654042 | 3654042 | 1 | |
| Insertion | Chr7 | 6490831 | 6490831 | 1 | |
| Insertion | Chr7 | 7927044 | 7927044 | 1 | |
| Insertion | Chr7 | 9513202 | 9513202 | 1 | |
| Insertion | Chr8 | 12071665 | 12071665 | 1 | |
| Insertion | Chr8 | 12245075 | 12245075 | 1 | |
| Insertion | Chr8 | 14434942 | 14434942 | 1 | LOC_Os08g23840 |
| Insertion | Chr8 | 15300347 | 15300348 | 2 | |
| Insertion | Chr8 | 18772592 | 18772604 | 13 | |
| Insertion | Chr8 | 19393944 | 19394005 | 62 | |
| Insertion | Chr8 | 22386121 | 22386121 | 1 | |
| Insertion | Chr8 | 2719368 | 2719369 | 2 | |
| Insertion | Chr8 | 4190770 | 4190770 | 1 | |
| Insertion | Chr8 | 6447731 | 6447731 | 1 | |
| Insertion | Chr8 | 7844749 | 7844749 | 1 | |
| Insertion | Chr9 | 1928159 | 1928159 | 1 | |
| Insertion | Chr9 | 583222 | 583223 | 2 | |
| Insertion | Chr9 | 6822950 | 6822950 | 1 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 3124201 | 3246605 | LOC_Os11g06670 |
| Inversion | Chr11 | 3124207 | 3246605 | LOC_Os11g06670 |
| Inversion | Chr2 | 4801609 | 4801681 | |
| Inversion | Chr9 | 13548782 | 13548946 | |
| Inversion | Chr7 | 23634914 | 23642874 | LOC_Os07g39460 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 9427622 | Chr1 | 35138890 | |
| Translocation | Chr5 | 10614492 | Chr1 | 14983725 | |
| Translocation | Chr12 | 19817004 | Chr7 | 23642860 | |
| Translocation | Chr12 | 19817163 | Chr7 | 23634888 |
