Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4455-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4455-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 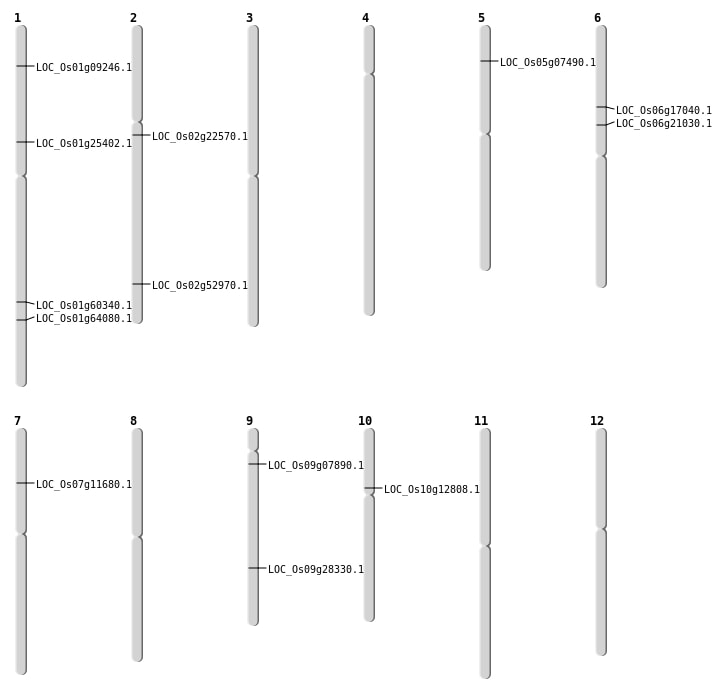
|
| Variant Information |
|---|
Single base substitutions: 51
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14371180 | C-T | HET | STOP_GAINED | LOC_Os01g25402 |
| SBS | Chr1 | 16373511 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 37228803 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g64080 |
| SBS | Chr1 | 38766484 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 42957774 | G-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 589355 | T-C | HET | INTRON | |
| SBS | Chr1 | 5931762 | A-G | HET | INTRON | |
| SBS | Chr1 | 8117322 | T-A | HET | INTRON | |
| SBS | Chr10 | 21828800 | A-C | HET | INTERGENIC | |
| SBS | Chr10 | 8109301 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 8109308 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 10628099 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 1145132 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 13576720 | G-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 25650243 | T-G | HOMO | SPLICE_SITE_REGION | |
| SBS | Chr11 | 9812976 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 13998496 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 19319945 | C-T | HET | INTRON | |
| SBS | Chr2 | 13457129 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g22570 |
| SBS | Chr2 | 17500435 | A-C | HET | INTRON | |
| SBS | Chr2 | 26260275 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 32407558 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 32407559 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g52970 |
| SBS | Chr2 | 32674295 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 9875409 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 10637413 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 17334041 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 19063180 | G-C | HET | INTRON | |
| SBS | Chr3 | 30412608 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 12871873 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 14369411 | A-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 18857907 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 29892861 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 32542191 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 33840511 | T-C | HET | INTRON | |
| SBS | Chr5 | 14737874 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 19448616 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 22647990 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 22647991 | C-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 28696161 | C-T | HOMO | INTRON | |
| SBS | Chr5 | 4001220 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g07490 |
| SBS | Chr5 | 9715068 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 8739357 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 21422251 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 6396602 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 12962956 | A-G | HOMO | INTRON | |
| SBS | Chr8 | 22515831 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 3808339 | T-C | HET | INTRON | |
| SBS | Chr9 | 17198216 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g28330 |
| SBS | Chr9 | 4011057 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g07890 |
| SBS | Chr9 | 713017 | T-C | HET | INTERGENIC |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 1975058 | 1975062 | 4 | |
| Deletion | Chr2 | 4468463 | 4468464 | 1 | |
| Deletion | Chr1 | 4666001 | 4685000 | 19000 | LOC_Os01g09246 |
| Deletion | Chr5 | 4737052 | 4737060 | 8 | |
| Deletion | Chr6 | 5391577 | 5391582 | 5 | |
| Deletion | Chr8 | 6176925 | 6176958 | 33 | |
| Deletion | Chr1 | 6540651 | 6540654 | 3 | |
| Deletion | Chr6 | 9436746 | 9436755 | 9 | |
| Deletion | Chr6 | 9881824 | 9881829 | 5 | LOC_Os06g17040 |
| Deletion | Chr7 | 10738885 | 10738886 | 1 | |
| Deletion | Chr6 | 12151612 | 12151613 | 1 | LOC_Os06g21030 |
| Deletion | Chr2 | 12155378 | 12155379 | 1 | |
| Deletion | Chr2 | 13414290 | 13414301 | 11 | |
| Deletion | Chr9 | 13652558 | 13652559 | 1 | |
| Deletion | Chr8 | 14086576 | 14086662 | 86 | |
| Deletion | Chr4 | 14495633 | 14495634 | 1 | |
| Deletion | Chr12 | 16307086 | 16307087 | 1 | |
| Deletion | Chr11 | 16655243 | 16655248 | 5 | |
| Deletion | Chr11 | 17164162 | 17164165 | 3 | |
| Deletion | Chr5 | 17496967 | 17496968 | 1 | |
| Deletion | Chr3 | 18917965 | 18917966 | 1 | |
| Deletion | Chr1 | 19534277 | 19534278 | 1 | |
| Deletion | Chr4 | 25220379 | 25220441 | 62 | |
| Deletion | Chr12 | 26151260 | 26151261 | 1 | |
| Deletion | Chr6 | 28495207 | 28495209 | 2 | |
| Deletion | Chr6 | 29723571 | 29723573 | 2 | |
| Deletion | Chr3 | 29966319 | 29966320 | 1 | |
| Deletion | Chr3 | 32937915 | 32937916 | 1 | |
| Deletion | Chr1 | 42158212 | 42158213 | 1 |
Insertions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 1106556 | 1106556 | 1 | |
| Insertion | Chr10 | 7159754 | 7159755 | 2 | LOC_Os10g12808 |
| Insertion | Chr12 | 18990627 | 18990628 | 2 | |
| Insertion | Chr12 | 8075404 | 8075405 | 2 | |
| Insertion | Chr2 | 31408220 | 31408220 | 1 | |
| Insertion | Chr2 | 9244220 | 9244223 | 4 | |
| Insertion | Chr3 | 23561629 | 23561629 | 1 | |
| Insertion | Chr3 | 30434909 | 30434910 | 2 | |
| Insertion | Chr3 | 35435881 | 35435881 | 1 | |
| Insertion | Chr4 | 17209076 | 17209076 | 1 | |
| Insertion | Chr4 | 2843144 | 2843145 | 2 | |
| Insertion | Chr5 | 14653158 | 14653158 | 1 | |
| Insertion | Chr6 | 22905987 | 22905987 | 1 | |
| Insertion | Chr6 | 3850197 | 3850197 | 1 | |
| Insertion | Chr7 | 6460545 | 6460545 | 1 | LOC_Os07g11680 |
| Insertion | Chr8 | 15859020 | 15859020 | 1 | |
| Insertion | Chr8 | 23437205 | 23437205 | 1 | |
| Insertion | Chr8 | 7266096 | 7266097 | 2 | |
| Insertion | Chr9 | 18576995 | 18576996 | 2 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 8645334 | 8645518 | |
| Inversion | Chr5 | 10754537 | 10820282 | |
| Inversion | Chr5 | 10754542 | 10820279 | |
| Inversion | Chr3 | 14726080 | 14726278 | |
| Inversion | Chr1 | 34902377 | 34902549 | LOC_Os01g60340 |
No Translocation
