Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4463-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4463-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 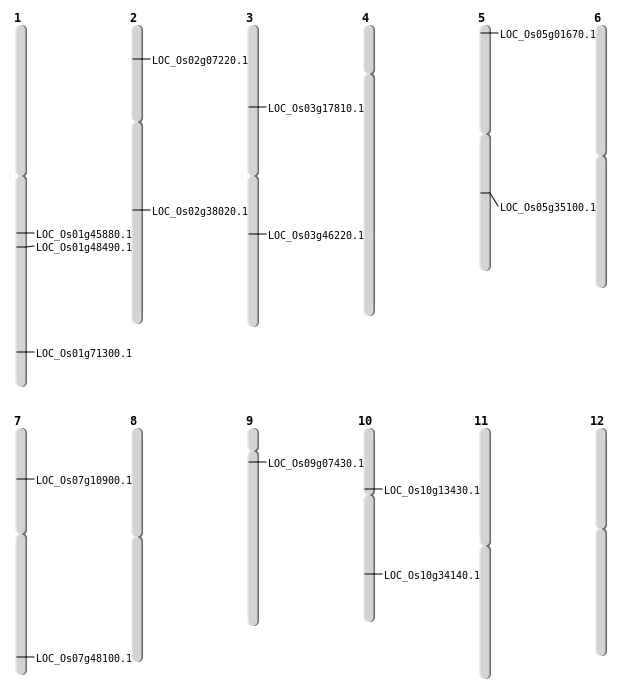
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14636635 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 23686248 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 27793872 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g48490 |
| SBS | Chr1 | 28227895 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 41271825 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g71300 |
| SBS | Chr10 | 5924660 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 7261599 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g13430 |
| SBS | Chr11 | 18745923 | T-A | HET | INTRON | |
| SBS | Chr11 | 23772745 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 7080693 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 7907136 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 9245966 | G-A | HET | INTRON | |
| SBS | Chr12 | 8250018 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 15648437 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 22974337 | T-A | HOMO | STOP_GAINED | LOC_Os02g38020 |
| SBS | Chr2 | 22974339 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g38020 |
| SBS | Chr2 | 22974342 | T-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g38020 |
| SBS | Chr3 | 17701094 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 27348174 | C-T | HET | INTRON | |
| SBS | Chr3 | 28646840 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 5776605 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 5776613 | A-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 9402730 | C-G | HET | START_GAINED | |
| SBS | Chr3 | 9916943 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g17810 |
| SBS | Chr4 | 10529898 | G-A | HET | INTRON | |
| SBS | Chr4 | 21129990 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 26176385 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 21634745 | A-C | HET | INTRON | |
| SBS | Chr5 | 398177 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 398178 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g01670 |
| SBS | Chr5 | 398179 | C-A | HET | STOP_GAINED | LOC_Os05g01670 |
| SBS | Chr6 | 23029823 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 883657 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 11166578 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 11962178 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 18957108 | G-A | HET | INTRON | |
| SBS | Chr7 | 26306351 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 28727472 | A-T | HET | STOP_GAINED | LOC_Os07g48100 |
| SBS | Chr7 | 5960930 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g10900 |
| SBS | Chr7 | 8682406 | C-T | HET | INTRON | |
| SBS | Chr7 | 8961431 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 9327097 | A-C | HET | INTERGENIC |
Deletions: 31
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 218661 | 218662 | 1 | |
| Deletion | Chr9 | 2437290 | 2437291 | 1 | |
| Deletion | Chr4 | 2564197 | 2564198 | 1 | |
| Deletion | Chr4 | 3080226 | 3080228 | 2 | |
| Deletion | Chr11 | 3795820 | 3795821 | 1 | |
| Deletion | Chr3 | 4932893 | 4932894 | 1 | |
| Deletion | Chr8 | 5046073 | 5046076 | 3 | |
| Deletion | Chr8 | 5786945 | 5786946 | 1 | |
| Deletion | Chr9 | 6658819 | 6658820 | 1 | |
| Deletion | Chr10 | 8223978 | 8223981 | 3 | |
| Deletion | Chr4 | 8635172 | 8635174 | 2 | |
| Deletion | Chr12 | 8680239 | 8680242 | 3 | |
| Deletion | Chr5 | 9904292 | 9904293 | 1 | |
| Deletion | Chr2 | 9964958 | 9964979 | 21 | |
| Deletion | Chr8 | 10790743 | 10790744 | 1 | |
| Deletion | Chr12 | 11689740 | 11689745 | 5 | |
| Deletion | Chr11 | 12104528 | 12104531 | 3 | |
| Deletion | Chr11 | 12108413 | 12108414 | 1 | |
| Deletion | Chr7 | 14259012 | 14259014 | 2 | |
| Deletion | Chr4 | 14976240 | 14976271 | 31 | |
| Deletion | Chr5 | 15367859 | 15367860 | 1 | |
| Deletion | Chr9 | 15670334 | 15670335 | 1 | |
| Deletion | Chr7 | 17256421 | 17256422 | 1 | |
| Deletion | Chr7 | 20797490 | 20797491 | 1 | |
| Deletion | Chr5 | 20839914 | 20839915 | 1 | LOC_Os05g35100 |
| Deletion | Chr7 | 24794232 | 24794233 | 1 | |
| Deletion | Chr1 | 26063515 | 26063516 | 1 | LOC_Os01g45880 |
| Deletion | Chr3 | 26131464 | 26131470 | 6 | LOC_Os03g46220 |
| Deletion | Chr2 | 26722355 | 26722376 | 21 | |
| Deletion | Chr3 | 26900011 | 26900027 | 16 | |
| Deletion | Chr6 | 29653116 | 29653117 | 1 |
Insertions: 16
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 18220085 | 18220275 | LOC_Os10g34140 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 3711240 | Chr2 | 7478966 | 2 |
| Translocation | Chr9 | 3712509 | Chr2 | 7478990 | LOC_Os09g07430 |
