Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4466-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4466-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 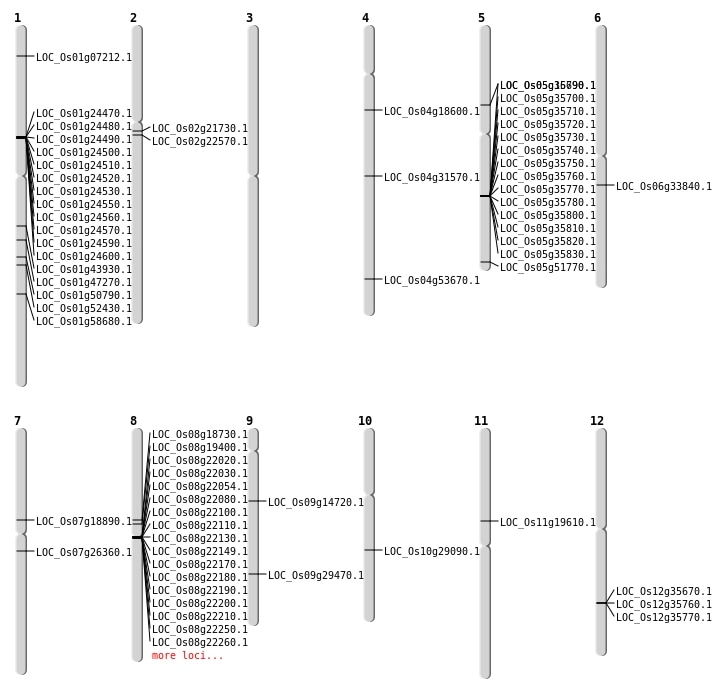
|
| Variant Information |
|---|
Single base substitutions: 48
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 19284483 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 23755260 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 27014758 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g47270 |
| SBS | Chr1 | 28805962 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 32516251 | T-C | HET | INTRON | |
| SBS | Chr1 | 33922001 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g58680 |
| SBS | Chr10 | 6756997 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 2519055 | C-G | HET | INTRON | |
| SBS | Chr12 | 11648761 | G-A | HET | INTRON | |
| SBS | Chr12 | 21694593 | C-G | HET | INTRON | |
| SBS | Chr12 | 2856680 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 4281938 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 683877 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 1118779 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 11845502 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr2 | 12985169 | T-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 12985170 | G-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 28933418 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 21553602 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 24920867 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 31618995 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 32268990 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 4773395 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 10275426 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g18600 |
| SBS | Chr4 | 18876328 | C-T | HOMO | STOP_GAINED | LOC_Os04g31570 |
| SBS | Chr4 | 29282402 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 31990410 | A-T | HET | STOP_GAINED | LOC_Os04g53670 |
| SBS | Chr5 | 10138637 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 10432801 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 13274141 | C-T | HOMO | INTRON | |
| SBS | Chr5 | 22202970 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 23168241 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 23612939 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 29237157 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 3903176 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 5453462 | C-G | HET | INTERGENIC | |
| SBS | Chr6 | 15751423 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 22462902 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 24734489 | G-A | HOMO | INTRON | |
| SBS | Chr6 | 27489317 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 31177988 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 5766826 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 11192707 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g18890 |
| SBS | Chr7 | 7639276 | T-A | HET | INTRON | |
| SBS | Chr7 | 9317960 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 11599518 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g19400 |
| SBS | Chr8 | 26591842 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 5782034 | A-T | HET | INTERGENIC |
Deletions: 49
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 206256 | 206257 | 1 | |
| Deletion | Chr1 | 706908 | 706925 | 17 | |
| Deletion | Chr7 | 733733 | 733734 | 1 | |
| Deletion | Chr10 | 1255092 | 1255093 | 1 | |
| Deletion | Chr6 | 1520509 | 1520510 | 1 | |
| Deletion | Chr1 | 1825272 | 1825351 | 79 | |
| Deletion | Chr12 | 2177958 | 2177959 | 1 | |
| Deletion | Chr4 | 3350476 | 3350499 | 23 | |
| Deletion | Chr1 | 3401709 | 3401717 | 8 | LOC_Os01g07212 |
| Deletion | Chr9 | 6687649 | 6687650 | 1 | |
| Deletion | Chr3 | 7263065 | 7263076 | 11 | |
| Deletion | Chr3 | 8989108 | 8989109 | 1 | |
| Deletion | Chr1 | 10150633 | 10150634 | 1 | |
| Deletion | Chr5 | 10649957 | 10649960 | 3 | |
| Deletion | Chr7 | 10968349 | 10968350 | 1 | |
| Deletion | Chr8 | 11181878 | 11181879 | 1 | LOC_Os08g18730 |
| Deletion | Chr3 | 11233465 | 11233478 | 13 | |
| Deletion | Chr7 | 11606100 | 11606105 | 5 | |
| Deletion | Chr12 | 11952720 | 11952722 | 2 | |
| Deletion | Chr3 | 12295487 | 12295488 | 1 | |
| Deletion | Chr5 | 12879547 | 12879552 | 5 | |
| Deletion | Chr2 | 12915939 | 12915943 | 4 | LOC_Os02g21730 |
| Deletion | Chr6 | 13224438 | 13224439 | 1 | |
| Deletion | Chr8 | 13292001 | 13462000 | 170000 | 21 |
| Deletion | Chr2 | 13455038 | 13455039 | 1 | LOC_Os02g22570 |
| Deletion | Chr8 | 13583682 | 13583685 | 3 | |
| Deletion | Chr1 | 13794001 | 13832000 | 38000 | 7 |
| Deletion | Chr1 | 13838001 | 13861000 | 23000 | 5 |
| Deletion | Chr8 | 14952447 | 14952448 | 1 | |
| Deletion | Chr8 | 16650401 | 16650412 | 11 | |
| Deletion | Chr2 | 16757935 | 16757937 | 2 | |
| Deletion | Chr12 | 16841767 | 16841770 | 3 | |
| Deletion | Chr4 | 16981874 | 16981877 | 3 | |
| Deletion | Chr6 | 19687794 | 19687799 | 5 | LOC_Os06g33840 |
| Deletion | Chr5 | 20452037 | 20452048 | 11 | |
| Deletion | Chr7 | 20486854 | 20486938 | 84 | |
| Deletion | Chr10 | 21079344 | 21079348 | 4 | |
| Deletion | Chr5 | 21198001 | 21264000 | 66000 | 14 |
| Deletion | Chr12 | 21773001 | 21789000 | 16000 | 2 |
| Deletion | Chr4 | 23257618 | 23257638 | 20 | |
| Deletion | Chr1 | 25174506 | 25174508 | 2 | LOC_Os01g43930 |
| Deletion | Chr6 | 25987424 | 25987462 | 38 | |
| Deletion | Chr4 | 26059208 | 26059209 | 1 | |
| Deletion | Chr11 | 26982412 | 26982413 | 1 | |
| Deletion | Chr1 | 27939720 | 27939722 | 2 | |
| Deletion | Chr1 | 29175998 | 29176040 | 42 | LOC_Os01g50790 |
| Deletion | Chr1 | 29640472 | 29640473 | 1 | |
| Deletion | Chr1 | 30126646 | 30126647 | 1 | LOC_Os01g52430 |
| Deletion | Chr1 | 39120498 | 39120516 | 18 |
Insertions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 19330590 | 19330590 | 1 | |
| Insertion | Chr1 | 20441901 | 20441901 | 1 | |
| Insertion | Chr1 | 28814598 | 28814600 | 3 | |
| Insertion | Chr1 | 30699386 | 30699386 | 1 | |
| Insertion | Chr10 | 2165624 | 2165625 | 2 | |
| Insertion | Chr11 | 27968595 | 27968595 | 1 | |
| Insertion | Chr11 | 7580134 | 7580135 | 2 | |
| Insertion | Chr11 | 7920716 | 7920717 | 2 | |
| Insertion | Chr12 | 23760671 | 23760742 | 72 | |
| Insertion | Chr3 | 26766884 | 26766899 | 16 | |
| Insertion | Chr4 | 10372738 | 10372740 | 3 | |
| Insertion | Chr4 | 1370481 | 1370481 | 1 | |
| Insertion | Chr4 | 13775750 | 13775750 | 1 | |
| Insertion | Chr4 | 23681155 | 23681155 | 1 | |
| Insertion | Chr5 | 11102261 | 11102262 | 2 | |
| Insertion | Chr5 | 12204265 | 12204265 | 1 | |
| Insertion | Chr5 | 21823814 | 21823815 | 2 | |
| Insertion | Chr5 | 3831408 | 3831408 | 1 | |
| Insertion | Chr6 | 10988216 | 10988216 | 1 | |
| Insertion | Chr6 | 11827213 | 11827213 | 1 | |
| Insertion | Chr6 | 21474906 | 21474906 | 1 | |
| Insertion | Chr6 | 30211584 | 30211594 | 11 | |
| Insertion | Chr7 | 22434571 | 22434572 | 2 | |
| Insertion | Chr7 | 24240108 | 24240186 | 79 | |
| Insertion | Chr7 | 24784941 | 24784990 | 50 | |
| Insertion | Chr8 | 13399250 | 13399251 | 2 | |
| Insertion | Chr8 | 14837365 | 14837365 | 1 | |
| Insertion | Chr9 | 13846489 | 13846490 | 2 | |
| Insertion | Chr9 | 17452420 | 17452420 | 1 | |
| Insertion | Chr9 | 8743942 | 8743942 | 1 | LOC_Os09g14720 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 21692582 | 21773080 | LOC_Os12g35670 |
Translocations: 10
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 7638010 | Chr2 | 3798617 | |
| Translocation | Chr7 | 9561138 | Chr5 | 12027106 | LOC_Os05g16790 |
| Translocation | Chr11 | 11305409 | Chr3 | 8275495 | LOC_Os11g19610 |
| Translocation | Chr10 | 15151887 | Chr7 | 1473203 | 2 |
| Translocation | Chr9 | 17923005 | Chr3 | 30851529 | LOC_Os09g29470 |
| Translocation | Chr9 | 17923171 | Chr3 | 30851494 | |
| Translocation | Chr11 | 18144085 | Chr2 | 35610329 | |
| Translocation | Chr7 | 22380316 | Chr3 | 18876768 | |
| Translocation | Chr7 | 22380317 | Chr3 | 18877003 | |
| Translocation | Chr5 | 29707326 | Chr2 | 18202509 | LOC_Os05g51770 |
