Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4478-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4478-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 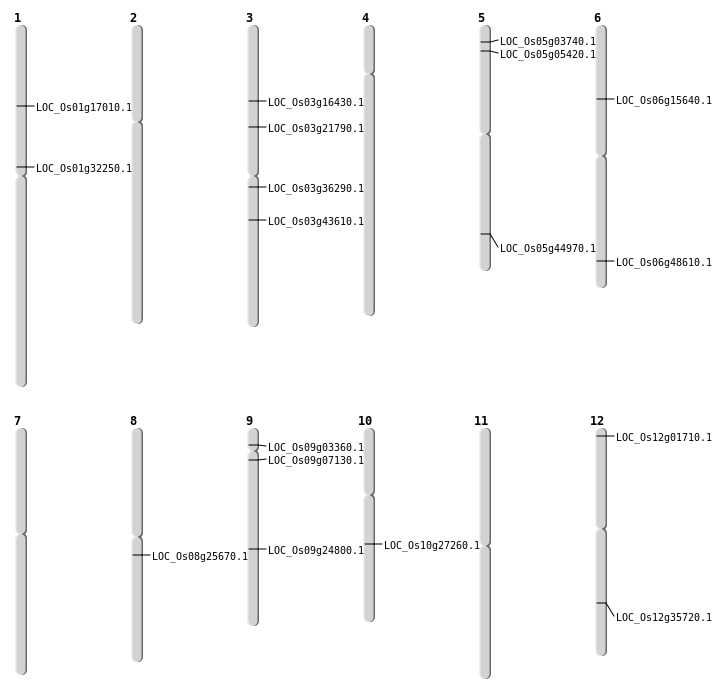
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr10 | 13401317 | C-A | HET | INTRON | |
| SBS | Chr10 | 15817455 | A-T | HET | INTRON | |
| SBS | Chr10 | 7908313 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 9169163 | T-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 12567658 | C-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 20875140 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 21739452 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g35720 |
| SBS | Chr12 | 9137400 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 13046400 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 17066123 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 17066127 | G-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 3710190 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 15839722 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 20131623 | G-A | HET | STOP_GAINED | LOC_Os03g36290 |
| SBS | Chr3 | 24374368 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g43610 |
| SBS | Chr3 | 26124347 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 6506541 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 1643881 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g03740 |
| SBS | Chr5 | 8664270 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 22862747 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 3117463 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 8860496 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g15640 |
| SBS | Chr7 | 11116990 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 14284216 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 23995263 | T-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 24982855 | C-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 7511671 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 13884079 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 15622051 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g25670 |
| SBS | Chr8 | 15685031 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 27269640 | G-T | HOMO | INTRON | |
| SBS | Chr8 | 3541689 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 11391953 | A-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 12405741 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 13274031 | C-T | HET | INTRON | |
| SBS | Chr9 | 1631142 | C-T | HET | STOP_GAINED | LOC_Os09g03360 |
| SBS | Chr9 | 21274108 | C-G | HET | INTRON |
Deletions: 37
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 422709 | 422723 | 14 | LOC_Os12g01710 |
| Deletion | Chr3 | 1106286 | 1106294 | 8 | |
| Deletion | Chr12 | 1672006 | 1672010 | 4 | |
| Deletion | Chr10 | 2462235 | 2462256 | 21 | |
| Deletion | Chr5 | 2690615 | 2690619 | 4 | LOC_Os05g05420 |
| Deletion | Chr3 | 2835577 | 2835600 | 23 | |
| Deletion | Chr9 | 4349164 | 4349165 | 1 | |
| Deletion | Chr12 | 5307125 | 5307126 | 1 | |
| Deletion | Chr8 | 5662476 | 5662487 | 11 | |
| Deletion | Chr8 | 8322114 | 8322115 | 1 | |
| Deletion | Chr1 | 9743965 | 9743971 | 6 | LOC_Os01g17010 |
| Deletion | Chr8 | 10226024 | 10226029 | 5 | |
| Deletion | Chr8 | 10561132 | 10561148 | 16 | |
| Deletion | Chr9 | 10979099 | 10979111 | 12 | |
| Deletion | Chr6 | 12493558 | 12493571 | 13 | |
| Deletion | Chr8 | 12651492 | 12651496 | 4 | |
| Deletion | Chr10 | 13865416 | 13865418 | 2 | |
| Deletion | Chr10 | 14381531 | 14381545 | 14 | LOC_Os10g27260 |
| Deletion | Chr12 | 15195711 | 15195781 | 70 | |
| Deletion | Chr3 | 15519273 | 15519275 | 2 | |
| Deletion | Chr1 | 15728310 | 15728352 | 42 | |
| Deletion | Chr12 | 15794524 | 15794526 | 2 | |
| Deletion | Chr9 | 16126355 | 16126356 | 1 | |
| Deletion | Chr12 | 16861037 | 16861038 | 1 | |
| Deletion | Chr3 | 17097648 | 17097652 | 4 | |
| Deletion | Chr1 | 17671004 | 17671005 | 1 | LOC_Os01g32250 |
| Deletion | Chr6 | 19913104 | 19913106 | 2 | |
| Deletion | Chr12 | 20895839 | 20895841 | 2 | |
| Deletion | Chr6 | 22062501 | 22062564 | 63 | |
| Deletion | Chr8 | 23297357 | 23297358 | 1 | |
| Deletion | Chr5 | 26144223 | 26144231 | 8 | LOC_Os05g44970 |
| Deletion | Chr5 | 26387280 | 26387286 | 6 | |
| Deletion | Chr11 | 28734953 | 28735034 | 81 | |
| Deletion | Chr5 | 29411022 | 29411023 | 1 | |
| Deletion | Chr1 | 34257260 | 34257263 | 3 | |
| Deletion | Chr4 | 35056978 | 35056994 | 16 | |
| Deletion | Chr1 | 41335477 | 41335478 | 1 |
Insertions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 36342653 | 36342653 | 1 | |
| Insertion | Chr11 | 18382735 | 18382736 | 2 | |
| Insertion | Chr12 | 23487966 | 23487975 | 10 | |
| Insertion | Chr2 | 28414957 | 28414958 | 2 | |
| Insertion | Chr2 | 8412118 | 8412119 | 2 | |
| Insertion | Chr3 | 12448369 | 12448369 | 1 | LOC_Os03g21790 |
| Insertion | Chr7 | 12473205 | 12473205 | 1 | |
| Insertion | Chr8 | 23489010 | 23489012 | 3 |
Inversions: 12
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 3362415 | 3362595 | |
| Inversion | Chr9 | 3483140 | 3483851 | LOC_Os09g07130 |
| Inversion | Chr6 | 7668658 | 7668773 | |
| Inversion | Chr3 | 9762216 | 9762381 | |
| Inversion | Chr9 | 14776349 | 15584871 | LOC_Os09g24800 |
| Inversion | Chr9 | 14776359 | 15584871 | LOC_Os09g24800 |
| Inversion | Chr6 | 16849707 | 16849820 | |
| Inversion | Chr3 | 19490145 | 19490346 | |
| Inversion | Chr6 | 29411149 | 29940887 | |
| Inversion | Chr6 | 29411208 | 29940897 | LOC_Os06g48610 |
| Inversion | Chr3 | 31421927 | 31722344 | |
| Inversion | Chr3 | 31421931 | 31722342 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 7704256 | Chr8 | 18415246 | |
| Translocation | Chr10 | 9066800 | Chr3 | 13497193 | LOC_Os03g16430 |
| Translocation | Chr11 | 27498163 | Chr7 | 25204012 |
