Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4481-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4481-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 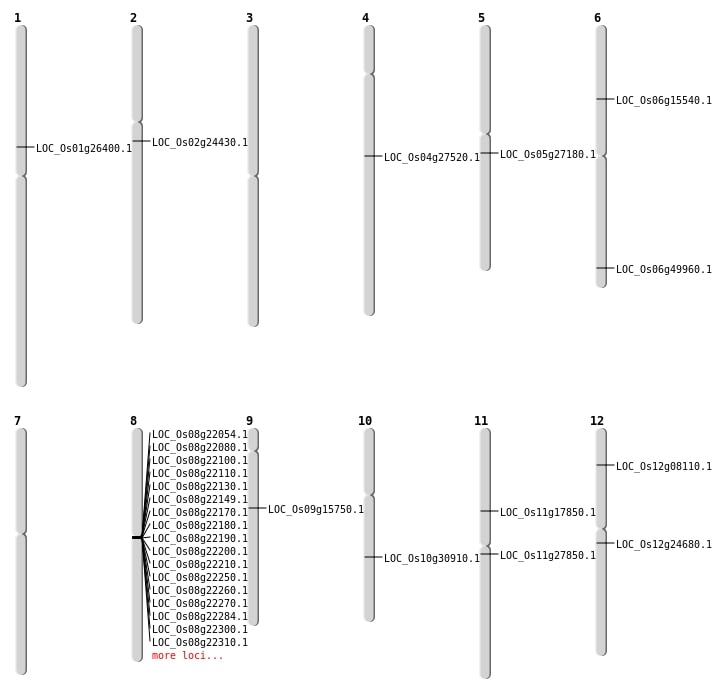
|
| Variant Information |
|---|
Single base substitutions: 44
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13732839 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 21100125 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 39480859 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 39816815 | C-T | HET | SPLICE_SITE_REGION | |
| SBS | Chr1 | 5212725 | T-C | HET | INTRON | |
| SBS | Chr1 | 5431653 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 16767995 | G-A | HET | SPLICE_SITE_REGION | |
| SBS | Chr10 | 19276578 | C-A | HET | INTRON | |
| SBS | Chr11 | 1413459 | G-T | HET | INTRON | |
| SBS | Chr11 | 16028557 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g27850 |
| SBS | Chr11 | 9993606 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g17850 |
| SBS | Chr12 | 13524521 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 15198987 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 2634355 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 2634356 | A-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 3995526 | A-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 8406212 | T-C | HET | INTRON | |
| SBS | Chr2 | 15666353 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 22258776 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 5905328 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 16269173 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g27520 |
| SBS | Chr4 | 26992106 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 32604522 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 4579776 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 14828528 | T-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 26553793 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 1107683 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 14526663 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 20416124 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 8811972 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g15540 |
| SBS | Chr7 | 1070125 | T-C | HET | INTRON | |
| SBS | Chr7 | 11679430 | G-T | HET | INTRON | |
| SBS | Chr7 | 15633013 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 25847373 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 27838357 | G-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 4004488 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 12320185 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 20414080 | G-A | HET | INTRON | |
| SBS | Chr8 | 20583970 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 8015298 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 15316200 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 16546335 | A-G | HOMO | INTRON | |
| SBS | Chr9 | 661415 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 9626785 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g15750 |
Deletions: 33
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 2710371 | 2710373 | 2 | |
| Deletion | Chr12 | 3489122 | 3489123 | 1 | |
| Deletion | Chr12 | 4133019 | 4133031 | 12 | LOC_Os12g08110 |
| Deletion | Chr1 | 4187174 | 4187175 | 1 | |
| Deletion | Chr12 | 4233833 | 4233835 | 2 | |
| Deletion | Chr1 | 4664582 | 4664583 | 1 | |
| Deletion | Chr1 | 6056431 | 6056432 | 1 | |
| Deletion | Chr5 | 6539473 | 6539474 | 1 | |
| Deletion | Chr12 | 7957828 | 7957842 | 14 | |
| Deletion | Chr8 | 8740457 | 8740460 | 3 | |
| Deletion | Chr11 | 9896224 | 9896286 | 62 | |
| Deletion | Chr8 | 13313001 | 13462000 | 149000 | 19 |
| Deletion | Chr12 | 14133670 | 14133703 | 33 | LOC_Os12g24680 |
| Deletion | Chr1 | 14984857 | 14984862 | 5 | LOC_Os01g26400 |
| Deletion | Chr4 | 15282126 | 15282129 | 3 | |
| Deletion | Chr5 | 15812061 | 15812062 | 1 | LOC_Os05g27180 |
| Deletion | Chr12 | 16995780 | 16995781 | 1 | |
| Deletion | Chr6 | 18938655 | 18938658 | 3 | |
| Deletion | Chr10 | 18999791 | 18999793 | 2 | |
| Deletion | Chr1 | 20580567 | 20580648 | 81 | |
| Deletion | Chr8 | 21000001 | 21055000 | 55000 | 10 |
| Deletion | Chr3 | 21722245 | 21722246 | 1 | |
| Deletion | Chr4 | 21745291 | 21745295 | 4 | |
| Deletion | Chr11 | 22372030 | 22372035 | 5 | |
| Deletion | Chr12 | 23217879 | 23217880 | 1 | |
| Deletion | Chr2 | 25127717 | 25127734 | 17 | |
| Deletion | Chr11 | 26024241 | 26024258 | 17 | |
| Deletion | Chr11 | 27030543 | 27030544 | 1 | |
| Deletion | Chr8 | 27210934 | 27210935 | 1 | |
| Deletion | Chr8 | 28207264 | 28207269 | 5 | |
| Deletion | Chr6 | 30254113 | 30254114 | 1 | LOC_Os06g49960 |
| Deletion | Chr1 | 31507863 | 31507869 | 6 | |
| Deletion | Chr1 | 39214415 | 39214416 | 1 |
Insertions: 11
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 11879326 | 12323915 | |
| Inversion | Chr2 | 14163763 | 14163958 | LOC_Os02g24430 |
| Inversion | Chr10 | 16125432 | 16667635 | LOC_Os10g30910 |
| Inversion | Chr6 | 17472425 | 17472583 | |
| Inversion | Chr8 | 20855629 | 21078802 | LOC_Os08g33430 |
| Inversion | Chr8 | 20855644 | 21078814 | LOC_Os08g33430 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 556778 | Chr2 | 34015809 |
