Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4498-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4498-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 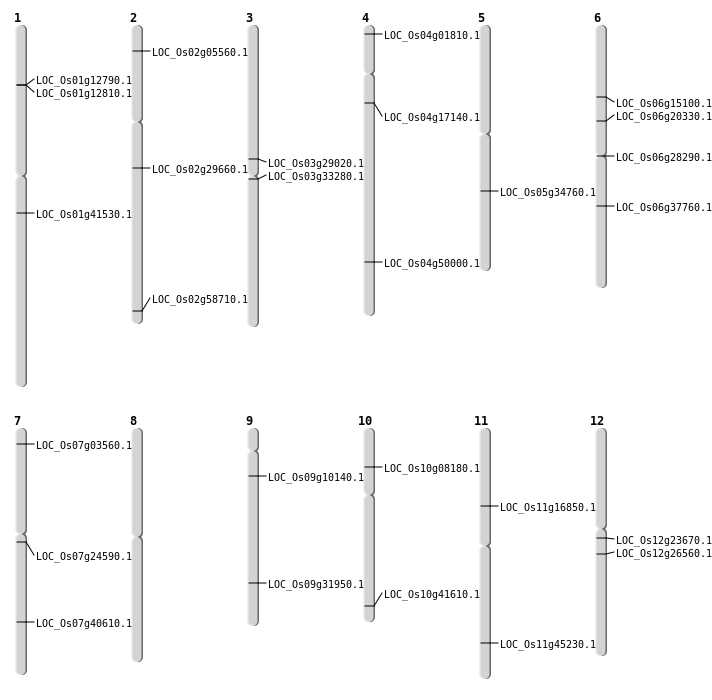
|
| Variant Information |
|---|
Single base substitutions: 69
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 19021600 | C-T | HET | ||
| SBS | Chr1 | 2973036 | T-C | HET | ||
| SBS | Chr1 | 7084677 | G-T | HET | LOC_Os01g12790 | |
| SBS | Chr1 | 7094440 | C-G | HET | LOC_Os01g12810 | |
| SBS | Chr1 | 7301673 | T-A | HET | ||
| SBS | Chr10 | 12386668 | C-T | HET | ||
| SBS | Chr10 | 14915385 | C-T | HET | ||
| SBS | Chr10 | 15008763 | T-C | HET | ||
| SBS | Chr10 | 17276395 | T-A | HET | ||
| SBS | Chr10 | 17536680 | C-G | HET | ||
| SBS | Chr10 | 19459676 | T-C | HOMO | ||
| SBS | Chr10 | 4424910 | A-T | HET | LOC_Os10g08180 | |
| SBS | Chr11 | 14988391 | G-T | HET | ||
| SBS | Chr11 | 1674935 | C-A | HET | ||
| SBS | Chr11 | 21153201 | C-T | HET | ||
| SBS | Chr11 | 26055311 | G-T | HOMO | ||
| SBS | Chr11 | 27372646 | G-T | HOMO | LOC_Os11g45230 | |
| SBS | Chr11 | 9354780 | C-A | HET | ||
| SBS | Chr11 | 9354781 | C-A | HET | LOC_Os11g16850 | |
| SBS | Chr12 | 13431970 | C-T | HET | LOC_Os12g23670 | |
| SBS | Chr12 | 17127385 | G-T | HOMO | ||
| SBS | Chr12 | 22246935 | A-G | HET | ||
| SBS | Chr12 | 23347860 | C-T | HET | ||
| SBS | Chr12 | 26093368 | G-T | HET | ||
| SBS | Chr12 | 26093369 | T-G | HET | ||
| SBS | Chr2 | 1111580 | G-A | HET | ||
| SBS | Chr2 | 12901312 | C-A | HET | ||
| SBS | Chr2 | 17658240 | G-C | HET | LOC_Os02g29660 | |
| SBS | Chr2 | 18769683 | G-T | HOMO | ||
| SBS | Chr2 | 19726600 | G-A | HET | ||
| SBS | Chr2 | 2696339 | T-G | HET | LOC_Os02g05560 | |
| SBS | Chr2 | 29740751 | A-T | HET | ||
| SBS | Chr2 | 35867711 | C-T | HOMO | LOC_Os02g58710 | |
| SBS | Chr3 | 12788125 | C-A | HOMO | ||
| SBS | Chr3 | 14024124 | C-T | HOMO | ||
| SBS | Chr3 | 19035129 | G-A | HET | LOC_Os03g33280 | |
| SBS | Chr3 | 25044447 | G-A | HET | ||
| SBS | Chr3 | 27465629 | G-A | HET | ||
| SBS | Chr3 | 27731676 | A-T | HET | ||
| SBS | Chr3 | 46039 | G-T | HET | ||
| SBS | Chr4 | 16302299 | A-T | HOMO | ||
| SBS | Chr4 | 1946912 | C-T | HOMO | ||
| SBS | Chr4 | 26455122 | A-T | HET | ||
| SBS | Chr4 | 29820569 | G-T | HET | LOC_Os04g50000 | |
| SBS | Chr4 | 3833704 | C-A | HET | ||
| SBS | Chr4 | 6546355 | G-T | HOMO | ||
| SBS | Chr5 | 10369529 | C-T | HET | ||
| SBS | Chr5 | 10705460 | C-T | HET | ||
| SBS | Chr5 | 15639445 | G-T | HET | ||
| SBS | Chr5 | 16122988 | G-A | HET | ||
| SBS | Chr5 | 20587555 | G-T | HET | ||
| SBS | Chr5 | 20587557 | A-T | HET | ||
| SBS | Chr5 | 25729610 | C-A | HET | ||
| SBS | Chr6 | 11682423 | T-C | HET | LOC_Os06g20330 | |
| SBS | Chr6 | 15758825 | A-G | HET | ||
| SBS | Chr6 | 22357251 | T-C | HET | LOC_Os06g37760 | |
| SBS | Chr6 | 7381769 | G-A | HET | ||
| SBS | Chr6 | 8329705 | C-T | HET | ||
| SBS | Chr6 | 8559674 | C-T | HET | LOC_Os06g15100 | |
| SBS | Chr7 | 13991998 | T-C | HET | LOC_Os07g24590 | |
| SBS | Chr7 | 14110879 | C-A | HET | ||
| SBS | Chr7 | 19603596 | A-T | HET | ||
| SBS | Chr7 | 5493733 | T-C | HET | ||
| SBS | Chr7 | 927310 | T-A | HET | ||
| SBS | Chr8 | 11520003 | G-C | HET | ||
| SBS | Chr8 | 13510509 | C-T | HET | ||
| SBS | Chr9 | 11058825 | C-G | HET | ||
| SBS | Chr9 | 19740806 | C-T | HOMO | ||
| SBS | Chr9 | 20104239 | G-A | HOMO |
Deletions: 48
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 2272854 | 2272854 | 1 | |
| Deletion | Chr2 | 3045226 | 3045233 | 8 | |
| Deletion | Chr7 | 3489595 | 3489595 | 1 | |
| Deletion | Chr9 | 4590610 | 4590610 | 1 | |
| Deletion | Chr10 | 4871521 | 4871522 | 2 | |
| Deletion | Chr8 | 5704069 | 5704070 | 2 | |
| Deletion | Chr8 | 6165890 | 6165890 | 1 | |
| Deletion | Chr8 | 6688550 | 6688550 | 1 | |
| Deletion | Chr7 | 8833360 | 8833360 | 1 | |
| Deletion | Chr4 | 9400140 | 9400147 | 8 | LOC_Os04g17140 |
| Deletion | Chr1 | 9786865 | 9786866 | 2 | |
| Deletion | Chr7 | 11451447 | 11451454 | 8 | |
| Deletion | Chr11 | 11733691 | 11733691 | 1 | |
| Deletion | Chr6 | 12434200 | 12434211 | 12 | |
| Deletion | Chr5 | 13239780 | 13239780 | 1 | |
| Deletion | Chr8 | 13361652 | 13361666 | 15 | |
| Deletion | Chr3 | 13468456 | 13468459 | 4 | |
| Deletion | Chr1 | 13625198 | 13625198 | 1 | |
| Deletion | Chr1 | 14254043 | 14254073 | 31 | |
| Deletion | Chr7 | 14931332 | 14931348 | 17 | |
| Deletion | Chr12 | 15525289 | 15525298 | 10 | LOC_Os12g26560 |
| Deletion | Chr8 | 15539798 | 15539801 | 4 | |
| Deletion | Chr11 | 16099491 | 16099502 | 12 | |
| Deletion | Chr3 | 16474817 | 16474832 | 16 | LOC_Os03g29020 |
| Deletion | Chr1 | 16628611 | 16628611 | 1 | |
| Deletion | Chr1 | 16894380 | 16894380 | 1 | |
| Deletion | Chr3 | 17945406 | 17945406 | 1 | |
| Deletion | Chr1 | 18277564 | 18277569 | 6 | |
| Deletion | Chr2 | 18769670 | 18769671 | 2 | |
| Deletion | Chr11 | 19790504 | 19790504 | 1 | |
| Deletion | Chr6 | 20323142 | 20323149 | 8 | |
| Deletion | Chr5 | 20603497 | 20603497 | 1 | |
| Deletion | Chr5 | 20625487 | 20625487 | 1 | LOC_Os05g34760 |
| Deletion | Chr5 | 22113896 | 22113897 | 2 | |
| Deletion | Chr2 | 22293367 | 22293367 | 1 | |
| Deletion | Chr10 | 22405187 | 22405188 | 2 | LOC_Os10g41610 |
| Deletion | Chr5 | 22567579 | 22567589 | 11 | |
| Deletion | Chr7 | 22798753 | 22798760 | 8 | |
| Deletion | Chr2 | 24253390 | 24253473 | 84 | |
| Deletion | Chr7 | 24335366 | 24335369 | 4 | LOC_Os07g40610 |
| Deletion | Chr6 | 24531108 | 24531109 | 2 | |
| Deletion | Chr12 | 25735822 | 25735822 | 1 | |
| Deletion | Chr11 | 25994498 | 25994498 | 1 | |
| Deletion | Chr2 | 26003675 | 26003676 | 2 | |
| Deletion | Chr7 | 26774183 | 26774200 | 18 | |
| Deletion | Chr2 | 28667087 | 28667096 | 10 | |
| Deletion | Chr3 | 28983778 | 28983778 | 1 | |
| Deletion | Chr3 | 31593514 | 31593514 | 1 |
Insertions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 15788693 | 15788693 | 1 | |
| Insertion | Chr1 | 30326319 | 30326319 | 1 | |
| Insertion | Chr1 | 32157225 | 32157225 | 1 | |
| Insertion | Chr1 | 34123379 | 34123379 | 1 | |
| Insertion | Chr10 | 18442930 | 18442930 | 1 | |
| Insertion | Chr10 | 7054922 | 7054922 | 1 | |
| Insertion | Chr11 | 12110389 | 12110436 | 48 | |
| Insertion | Chr11 | 12172742 | 12172819 | 78 | |
| Insertion | Chr12 | 19334771 | 19334771 | 1 | |
| Insertion | Chr12 | 25527266 | 25527267 | 2 | |
| Insertion | Chr12 | 6962567 | 6962573 | 7 | |
| Insertion | Chr12 | 905434 | 905435 | 2 | |
| Insertion | Chr12 | 9829199 | 9829199 | 1 | |
| Insertion | Chr2 | 12118827 | 12118913 | 87 | |
| Insertion | Chr2 | 18198963 | 18198965 | 3 | |
| Insertion | Chr2 | 22771892 | 22771914 | 23 | |
| Insertion | Chr3 | 14795103 | 14795103 | 1 | |
| Insertion | Chr4 | 27972062 | 27972075 | 14 | |
| Insertion | Chr4 | 3977257 | 3977279 | 23 | |
| Insertion | Chr4 | 9035808 | 9035808 | 1 | |
| Insertion | Chr5 | 1696042 | 1696042 | 1 | |
| Insertion | Chr6 | 3779538 | 3779538 | 1 | |
| Insertion | Chr6 | 8546516 | 8546516 | 1 | |
| Insertion | Chr7 | 14148063 | 14148112 | 50 | |
| Insertion | Chr7 | 18184723 | 18184723 | 1 | |
| Insertion | Chr7 | 2417384 | 2417384 | 1 | |
| Insertion | Chr7 | 26765375 | 26765375 | 1 | |
| Insertion | Chr8 | 11044147 | 11044190 | 44 | |
| Insertion | Chr8 | 19945355 | 19945361 | 7 | |
| Insertion | Chr8 | 20164254 | 20164359 | 106 | |
| Insertion | Chr9 | 14134995 | 14135005 | 11 | |
| Insertion | Chr9 | 3178230 | 3178231 | 2 | |
| Insertion | Chr9 | 5517624 | 5517624 | 1 | LOC_Os09g10140 |
| Insertion | Chr9 | 7385407 | 7385408 | 2 |
Inversions: 6
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 16998603 | Chr6 | 21004596 | |
| Translocation | Chr11 | 18846692 | Chr4 | 16025045 | |
| Translocation | Chr2 | 20476679 | Chr1 | 23511189 | LOC_Os01g41530 |
| Translocation | Chr2 | 20476688 | Chr1 | 24042911 |
