Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4545-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4545-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 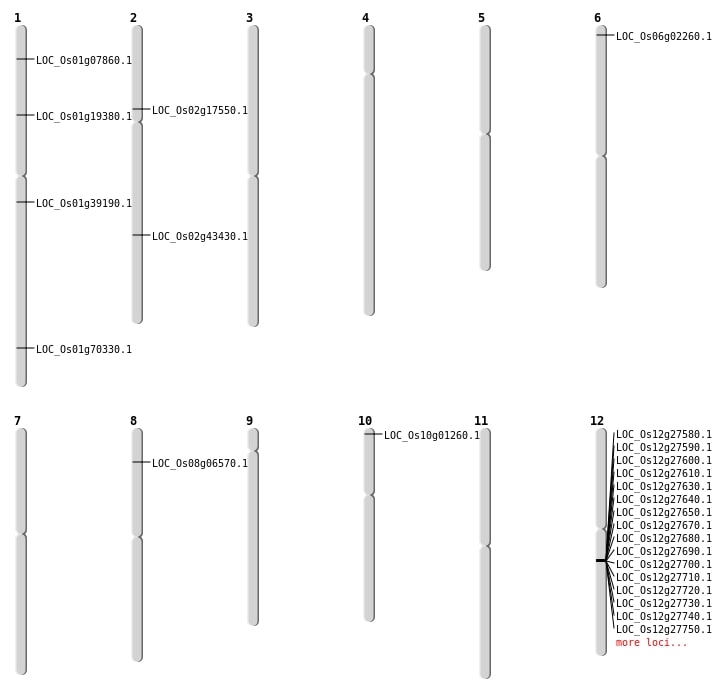
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 22046291 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g39190 |
| SBS | Chr1 | 296547 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 296548 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 37697946 | T-A | HOMO | INTRON | |
| SBS | Chr1 | 6555449 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 170814 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g01260 |
| SBS | Chr10 | 1890395 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr10 | 7796689 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 21917031 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 5601415 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 10102526 | G-T | HET | STOP_GAINED | LOC_Os02g17550 |
| SBS | Chr2 | 11008110 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 12673635 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 15965125 | A-G | HET | INTRON | |
| SBS | Chr2 | 25855244 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 26228986 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 11796980 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 14713910 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 25821494 | G-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 26843121 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 27183124 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 17434897 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 24112106 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 17625422 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 18231864 | T-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 4901032 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 725413 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g02260 |
| SBS | Chr7 | 2858565 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 15323845 | T-A | HET | INTRON | |
| SBS | Chr8 | 3708670 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g06570 |
| SBS | Chr8 | 6898529 | G-A | HET | INTERGENIC |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 2960840 | 2960842 | 2 | |
| Deletion | Chr1 | 3784093 | 3784094 | 1 | LOC_Os01g07860 |
| Deletion | Chr4 | 4187104 | 4187108 | 4 | |
| Deletion | Chr7 | 4948604 | 4948605 | 1 | |
| Deletion | Chr10 | 7851291 | 7851292 | 1 | |
| Deletion | Chr4 | 8438412 | 8438414 | 2 | |
| Deletion | Chr2 | 9469851 | 9469860 | 9 | |
| Deletion | Chr1 | 10884004 | 10884006 | 2 | |
| Deletion | Chr1 | 10965276 | 10965277 | 1 | LOC_Os01g19380 |
| Deletion | Chr5 | 12136299 | 12136301 | 2 | |
| Deletion | Chr1 | 12473482 | 12473483 | 1 | |
| Deletion | Chr5 | 13084369 | 13084370 | 1 | |
| Deletion | Chr5 | 13503610 | 13503615 | 5 | |
| Deletion | Chr10 | 14037576 | 14037579 | 3 | |
| Deletion | Chr7 | 14787853 | 14787854 | 1 | |
| Deletion | Chr6 | 15824475 | 15824479 | 4 | |
| Deletion | Chr12 | 16229001 | 16269000 | 40000 | 5 |
| Deletion | Chr12 | 16273001 | 16439000 | 166000 | 24 |
| Deletion | Chr12 | 16447001 | 16603000 | 156000 | 20 |
| Deletion | Chr12 | 16650001 | 16718000 | 68000 | 8 |
| Deletion | Chr7 | 22012956 | 22012957 | 1 | |
| Deletion | Chr7 | 22157258 | 22157262 | 4 | |
| Deletion | Chr2 | 23445168 | 23445171 | 3 | |
| Deletion | Chr2 | 24447641 | 24447661 | 20 | |
| Deletion | Chr2 | 26212524 | 26212539 | 15 | LOC_Os02g43430 |
| Deletion | Chr6 | 28755479 | 28755480 | 1 | |
| Deletion | Chr4 | 29264682 | 29264684 | 2 | |
| Deletion | Chr5 | 29356452 | 29356459 | 7 | |
| Deletion | Chr1 | 42224612 | 42224613 | 1 |
Insertions: 16
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 2543839 | 2544048 | |
| Inversion | Chr7 | 11237081 | 11237222 | |
| Inversion | Chr2 | 27127344 | 27127509 | |
| Inversion | Chr1 | 40741130 | 40741284 | LOC_Os01g70330 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 16718232 | Chr4 | 2584620 |
