Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4556-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4556-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 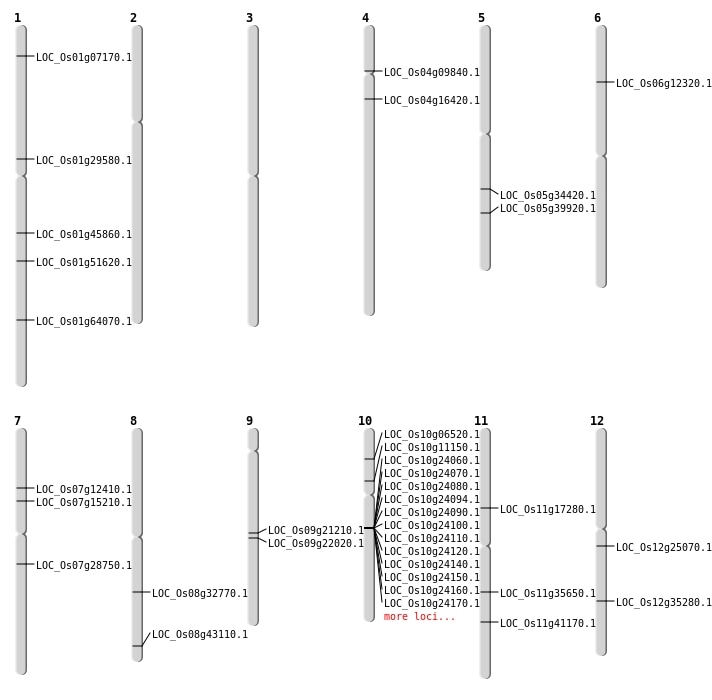
|
| Variant Information |
|---|
Single base substitutions: 48
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16578160 | C-T | HET | LOC_Os01g29580 | |
| SBS | Chr1 | 20651357 | A-T | HET | ||
| SBS | Chr1 | 26044888 | C-T | HET | LOC_Os01g45860 | |
| SBS | Chr10 | 12948419 | A-T | HOMO | ||
| SBS | Chr11 | 13115791 | C-G | HET | ||
| SBS | Chr11 | 13862668 | T-A | HET | ||
| SBS | Chr11 | 15294931 | C-T | HET | ||
| SBS | Chr11 | 16028906 | G-A | HET | ||
| SBS | Chr11 | 20201467 | G-A | HET | ||
| SBS | Chr11 | 20900871 | C-T | HET | LOC_Os11g35650 | |
| SBS | Chr11 | 21267725 | C-T | HET | ||
| SBS | Chr11 | 28349967 | G-A | HET | ||
| SBS | Chr11 | 516610 | G-T | HET | ||
| SBS | Chr11 | 516611 | C-T | HET | ||
| SBS | Chr11 | 516612 | G-T | HET | ||
| SBS | Chr11 | 988107 | C-T | HET | ||
| SBS | Chr12 | 14402641 | C-A | HET | LOC_Os12g25070 | |
| SBS | Chr12 | 16837224 | C-A | HET | ||
| SBS | Chr12 | 21438615 | G-A | HET | LOC_Os12g35280 | |
| SBS | Chr12 | 7853097 | A-G | HET | ||
| SBS | Chr2 | 31992514 | T-A | HOMO | ||
| SBS | Chr2 | 34254887 | C-T | HET | ||
| SBS | Chr3 | 1061090 | T-A | HET | ||
| SBS | Chr3 | 10747950 | G-A | HET | ||
| SBS | Chr3 | 12399158 | T-A | HOMO | ||
| SBS | Chr3 | 2493091 | T-G | HET | ||
| SBS | Chr3 | 33154424 | C-A | HET | ||
| SBS | Chr4 | 12022318 | C-T | HET | ||
| SBS | Chr4 | 32024781 | T-C | HOMO | ||
| SBS | Chr4 | 5769879 | A-G | HET | ||
| SBS | Chr5 | 19318655 | G-T | HET | ||
| SBS | Chr5 | 19318656 | G-T | HET | ||
| SBS | Chr5 | 6439560 | G-A | HET | ||
| SBS | Chr6 | 13889971 | T-C | HOMO | ||
| SBS | Chr6 | 3092895 | T-G | HET | ||
| SBS | Chr7 | 18008974 | G-T | HET | ||
| SBS | Chr7 | 27494258 | A-G | HET | ||
| SBS | Chr7 | 5501415 | C-T | HOMO | ||
| SBS | Chr7 | 8133094 | T-C | HOMO | ||
| SBS | Chr8 | 10334680 | A-G | HET | ||
| SBS | Chr8 | 3571655 | A-G | HOMO | ||
| SBS | Chr9 | 11716418 | T-C | HET | ||
| SBS | Chr9 | 13336715 | G-A | HET | LOC_Os09g22020 | |
| SBS | Chr9 | 1821177 | A-G | HOMO | ||
| SBS | Chr9 | 18223848 | C-A | HOMO | ||
| SBS | Chr9 | 3745189 | C-T | HOMO | ||
| SBS | Chr9 | 4815938 | C-A | HET | ||
| SBS | Chr9 | 8308264 | A-T | HET |
Deletions: 37
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 2038538 | 2038538 | 1 | |
| Deletion | Chr10 | 3349462 | 3349730 | 269 | LOC_Os10g06520 |
| Deletion | Chr5 | 5413959 | 5413959 | 1 | |
| Deletion | Chr7 | 5623845 | 5623845 | 1 | |
| Deletion | Chr6 | 6702241 | 6702253 | 13 | |
| Deletion | Chr7 | 7079032 | 7079032 | 1 | LOC_Os07g12410 |
| Deletion | Chr7 | 7610786 | 7610788 | 3 | |
| Deletion | Chr2 | 8422761 | 8422767 | 7 | |
| Deletion | Chr4 | 8926402 | 8926406 | 5 | LOC_Os04g16420 |
| Deletion | Chr2 | 9812718 | 9812718 | 1 | |
| Deletion | Chr3 | 10074159 | 10074169 | 11 | |
| Deletion | Chr10 | 11107372 | 11107374 | 3 | |
| Deletion | Chr12 | 11141520 | 11141603 | 84 | |
| Deletion | Chr10 | 12325001 | 12534000 | 209000 | 38 |
| Deletion | Chr10 | 12560991 | 12560994 | 4 | |
| Deletion | Chr6 | 13465604 | 13465604 | 1 | |
| Deletion | Chr6 | 14743939 | 14743940 | 2 | |
| Deletion | Chr4 | 17335635 | 17335727 | 93 | |
| Deletion | Chr4 | 17527131 | 17527174 | 44 | |
| Deletion | Chr8 | 18080317 | 18080322 | 6 | |
| Deletion | Chr10 | 18591378 | 18591379 | 2 | |
| Deletion | Chr4 | 18699659 | 18699660 | 2 | |
| Deletion | Chr3 | 19030045 | 19030046 | 2 | |
| Deletion | Chr5 | 20397125 | 20397216 | 92 | LOC_Os05g34420 |
| Deletion | Chr7 | 20899762 | 20899765 | 4 | |
| Deletion | Chr10 | 22184576 | 22184576 | 1 | |
| Deletion | Chr6 | 22730348 | 22730438 | 91 | |
| Deletion | Chr2 | 22766655 | 22766696 | 42 | |
| Deletion | Chr12 | 22891456 | 22891456 | 1 | |
| Deletion | Chr5 | 23457499 | 23457511 | 13 | LOC_Os05g39920 |
| Deletion | Chr11 | 24671133 | 24671133 | 1 | LOC_Os11g41170 |
| Deletion | Chr5 | 25368749 | 25368749 | 1 | LOC_Os05g43630 |
| Deletion | Chr12 | 26529184 | 26529187 | 4 | |
| Deletion | Chr7 | 28191176 | 28191182 | 7 | |
| Deletion | Chr6 | 29273253 | 29273253 | 1 | |
| Deletion | Chr6 | 29849962 | 29849971 | 10 | |
| Deletion | Chr5 | 29942014 | 29942015 | 2 |
Insertions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 37213454 | 37213454 | 1 | LOC_Os01g64070 |
| Insertion | Chr10 | 22612322 | 22612322 | 1 | |
| Insertion | Chr10 | 22719832 | 22719847 | 16 | |
| Insertion | Chr10 | 4627326 | 4627329 | 4 | |
| Insertion | Chr11 | 16926333 | 16926408 | 76 | |
| Insertion | Chr2 | 19910409 | 19910409 | 1 | |
| Insertion | Chr2 | 24759160 | 24759160 | 1 | |
| Insertion | Chr4 | 18682702 | 18682706 | 5 | |
| Insertion | Chr4 | 33664240 | 33664240 | 1 | |
| Insertion | Chr4 | 5284154 | 5284154 | 1 | LOC_Os04g09840 |
| Insertion | Chr6 | 20871572 | 20871654 | 83 | |
| Insertion | Chr6 | 23276720 | 23276720 | 1 | |
| Insertion | Chr6 | 8507675 | 8507677 | 3 | |
| Insertion | Chr7 | 11803365 | 11803365 | 1 | |
| Insertion | Chr7 | 16828819 | 16828819 | 1 | LOC_Os07g28750 |
| Insertion | Chr7 | 8766718 | 8766718 | 1 | LOC_Os07g15210 |
| Insertion | Chr8 | 12104566 | 12104585 | 20 | |
| Insertion | Chr8 | 27255845 | 27255845 | 1 | LOC_Os08g43110 |
| Insertion | Chr8 | 328363 | 328363 | 1 |
Inversions: 6
No Translocation
