Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN456-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN456-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 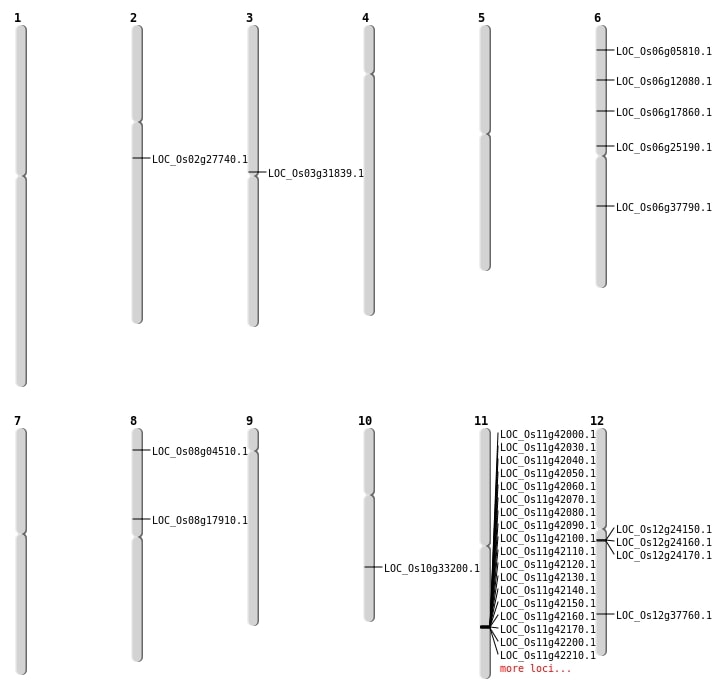
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16224498 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 28566540 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 30479227 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 36646900 | T-C | HOMO | UTR_3_PRIME | |
| SBS | Chr1 | 40685653 | T-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 16558249 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 19772327 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 20120036 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 22785591 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 5486005 | T-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 26532652 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 28344190 | T-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g47130 |
| SBS | Chr12 | 14554936 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 17719998 | A-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 19509480 | G-T | HOMO | INTRON | |
| SBS | Chr12 | 27166873 | G-C | HET | UTR_3_PRIME | |
| SBS | Chr12 | 5360423 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 8954890 | G-T | HOMO | INTRON | |
| SBS | Chr2 | 15240744 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 302620 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 34394612 | T-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 34885316 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 10792584 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 12122074 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 24957348 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 24957349 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 25260359 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 31189258 | T-G | HET | INTRON | |
| SBS | Chr5 | 16500579 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 17276253 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 8766577 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 2646026 | G-A | HET | STOP_GAINED | LOC_Os06g05810 |
| SBS | Chr6 | 8644248 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 14026266 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 20823905 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 10987891 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g17910 |
| SBS | Chr8 | 12594922 | G-C | HET | INTERGENIC | |
| SBS | Chr8 | 20203369 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 2224660 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g04510 |
| SBS | Chr8 | 25324692 | C-T | HET | INTRON | |
| SBS | Chr9 | 10051383 | T-C | HET | INTRON | |
| SBS | Chr9 | 2157199 | T-G | HET | SYNONYMOUS_CODING |
Deletions: 31
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 128203 | 128204 | 1 | |
| Deletion | Chr4 | 1663426 | 1663429 | 3 | |
| Deletion | Chr1 | 4383867 | 4383875 | 8 | |
| Deletion | Chr4 | 7870928 | 7870934 | 6 | |
| Deletion | Chr9 | 12089095 | 12089097 | 2 | |
| Deletion | Chr9 | 12979740 | 12979742 | 2 | |
| Deletion | Chr9 | 13358519 | 13358523 | 4 | |
| Deletion | Chr12 | 13744001 | 13769000 | 25000 | 3 |
| Deletion | Chr1 | 13951963 | 13951964 | 1 | |
| Deletion | Chr2 | 15158254 | 15158285 | 31 | |
| Deletion | Chr8 | 16067469 | 16067470 | 1 | |
| Deletion | Chr2 | 16212042 | 16212053 | 11 | |
| Deletion | Chr8 | 17218170 | 17218178 | 8 | |
| Deletion | Chr10 | 18826146 | 18826149 | 3 | |
| Deletion | Chr8 | 19213259 | 19213265 | 6 | |
| Deletion | Chr4 | 20574010 | 20574011 | 1 | |
| Deletion | Chr3 | 20854070 | 20854073 | 3 | |
| Deletion | Chr6 | 22364871 | 22364883 | 12 | LOC_Os06g37790 |
| Deletion | Chr3 | 22465835 | 22465846 | 11 | |
| Deletion | Chr6 | 23016897 | 23016898 | 1 | |
| Deletion | Chr2 | 24844454 | 24844460 | 6 | |
| Deletion | Chr11 | 25276001 | 25406000 | 130000 | 15 |
| Deletion | Chr11 | 25417001 | 25594000 | 177000 | 30 |
| Deletion | Chr7 | 28438382 | 28438383 | 1 | |
| Deletion | Chr4 | 30333426 | 30333431 | 5 | |
| Deletion | Chr1 | 30932584 | 30932589 | 5 | |
| Deletion | Chr1 | 31156980 | 31156984 | 4 | |
| Deletion | Chr2 | 32555287 | 32555301 | 14 | |
| Deletion | Chr2 | 34973929 | 34973933 | 4 | |
| Deletion | Chr2 | 35861889 | 35861890 | 1 | |
| Deletion | Chr1 | 38407838 | 38407851 | 13 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 25813217 | 25813220 | 4 | |
| Insertion | Chr4 | 8411775 | 8411775 | 1 | |
| Insertion | Chr5 | 2567681 | 2567682 | 2 | |
| Insertion | Chr8 | 20502371 | 20502371 | 1 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 677330 | 13216940 | |
| Inversion | Chr11 | 5603295 | 25593863 | LOC_Os11g42500 |
| Inversion | Chr11 | 5603538 | 25261252 | LOC_Os11g42000 |
| Inversion | Chr6 | 6451422 | 6473547 | LOC_Os06g12080 |
| Inversion | Chr6 | 10366408 | 14738921 | 2 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 16229027 | Chr1 | 42158279 | |
| Translocation | Chr11 | 16997659 | Chr2 | 474828 | |
| Translocation | Chr7 | 21204845 | Chr6 | 8411333 | |
| Translocation | Chr12 | 23184050 | Chr10 | 17400433 | 2 |
| Translocation | Chr12 | 26150514 | Chr2 | 16427021 | LOC_Os02g27740 |
| Translocation | Chr11 | 28162517 | Chr5 | 14142168 | LOC_Os11g46880 |
| Translocation | Chr8 | 28319003 | Chr5 | 5289576 | |
| Translocation | Chr4 | 34006298 | Chr3 | 18216873 | LOC_Os03g31839 |
