Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4560-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4560-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 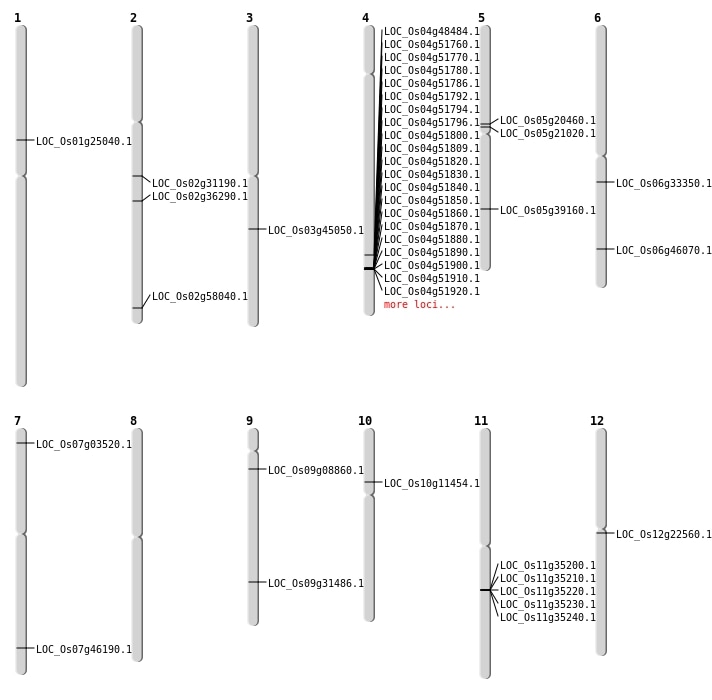
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14125077 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g25040 |
| SBS | Chr1 | 14583727 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 22390501 | A-C | HET | INTERGENIC | |
| SBS | Chr10 | 20294995 | G-C | HET | INTRON | |
| SBS | Chr10 | 4728911 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 6340163 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g11454 |
| SBS | Chr11 | 10291499 | G-T | HET | INTRON | |
| SBS | Chr11 | 13507607 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 22536539 | C-G | HET | UTR_3_PRIME | |
| SBS | Chr11 | 23305864 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 12735796 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g22560 |
| SBS | Chr12 | 13770374 | C-A | HET | INTRON | |
| SBS | Chr12 | 25083703 | G-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 5579252 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 6603085 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 24207819 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 25424579 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g45050 |
| SBS | Chr4 | 1536115 | G-T | HOMO | INTRON | |
| SBS | Chr4 | 35190251 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 6641555 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 12009050 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g20460 |
| SBS | Chr5 | 15659988 | T-A | HOMO | INTRON | |
| SBS | Chr5 | 21183533 | A-G | HOMO | INTRON | |
| SBS | Chr5 | 22966648 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g39160 |
| SBS | Chr5 | 29688059 | T-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 28545278 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 919068 | T-C | HET | INTRON | |
| SBS | Chr7 | 27571390 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g46190 |
| SBS | Chr7 | 2977444 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 6588263 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 20417932 | A-C | HET | SPLICE_SITE_REGION | |
| SBS | Chr8 | 2655604 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 3545034 | C-A | HET | SYNONYMOUS_CODING |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 2999553 | 2999554 | 1 | |
| Deletion | Chr5 | 4474970 | 4474972 | 2 | |
| Deletion | Chr9 | 4682623 | 4682641 | 18 | LOC_Os09g08860 |
| Deletion | Chr9 | 5805533 | 5805534 | 1 | |
| Deletion | Chr9 | 5891285 | 5891286 | 1 | |
| Deletion | Chr8 | 6771211 | 6771212 | 1 | |
| Deletion | Chr9 | 8253070 | 8253071 | 1 | |
| Deletion | Chr8 | 8739977 | 8739978 | 1 | |
| Deletion | Chr5 | 9005281 | 9005286 | 5 | |
| Deletion | Chr5 | 10342407 | 10342413 | 6 | |
| Deletion | Chr6 | 11354075 | 11354076 | 1 | |
| Deletion | Chr11 | 14972928 | 14972929 | 1 | |
| Deletion | Chr7 | 15083740 | 15083768 | 28 | |
| Deletion | Chr9 | 18988943 | 18988955 | 12 | LOC_Os09g31486 |
| Deletion | Chr6 | 19429695 | 19429706 | 11 | LOC_Os06g33350 |
| Deletion | Chr11 | 20039687 | 20039689 | 2 | |
| Deletion | Chr11 | 20635001 | 20663000 | 28000 | 5 |
| Deletion | Chr1 | 22767073 | 22767075 | 2 | |
| Deletion | Chr12 | 24971645 | 24971646 | 1 | |
| Deletion | Chr4 | 26414689 | 26414690 | 1 | |
| Deletion | Chr1 | 26582459 | 26582461 | 2 | |
| Deletion | Chr6 | 27934696 | 27934701 | 5 | LOC_Os06g46070 |
| Deletion | Chr4 | 30671001 | 30890000 | 219000 | 29 |
| Deletion | Chr4 | 30895001 | 31206000 | 311000 | 41 |
| Deletion | Chr3 | 34655914 | 34655915 | 1 | |
| Deletion | Chr4 | 35038835 | 35038842 | 7 |
Insertions: 17
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 35491259 | 35491264 | 6 | |
| Insertion | Chr10 | 14271920 | 14271920 | 1 | |
| Insertion | Chr12 | 11539103 | 11539103 | 1 | |
| Insertion | Chr12 | 5511207 | 5511214 | 8 | |
| Insertion | Chr12 | 7850625 | 7850636 | 12 | |
| Insertion | Chr2 | 12254823 | 12254824 | 2 | |
| Insertion | Chr2 | 18680854 | 18680854 | 1 | LOC_Os02g31190 |
| Insertion | Chr2 | 21913119 | 21913119 | 1 | LOC_Os02g36290 |
| Insertion | Chr4 | 2658320 | 2658320 | 1 | |
| Insertion | Chr4 | 2767186 | 2767188 | 3 | |
| Insertion | Chr5 | 12367664 | 12367666 | 3 | LOC_Os05g21020 |
| Insertion | Chr5 | 15388851 | 15388852 | 2 | |
| Insertion | Chr5 | 8165201 | 8165216 | 16 | |
| Insertion | Chr6 | 1784702 | 1784702 | 1 | |
| Insertion | Chr6 | 8170445 | 8170450 | 6 | |
| Insertion | Chr6 | 850515 | 850515 | 1 | |
| Insertion | Chr7 | 1406972 | 1406972 | 1 | LOC_Os07g03520 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 24278524 | 24525330 | |
| Inversion | Chr5 | 24278527 | 24525334 | |
| Inversion | Chr2 | 34963256 | 35535811 | LOC_Os02g58040 |
| Inversion | Chr2 | 34963294 | 35535852 | LOC_Os02g58040 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 28904022 | Chr1 | 26271178 | LOC_Os04g48484 |
| Translocation | Chr4 | 28904025 | Chr1 | 26271292 | LOC_Os04g48484 |
