Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4593-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4593-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 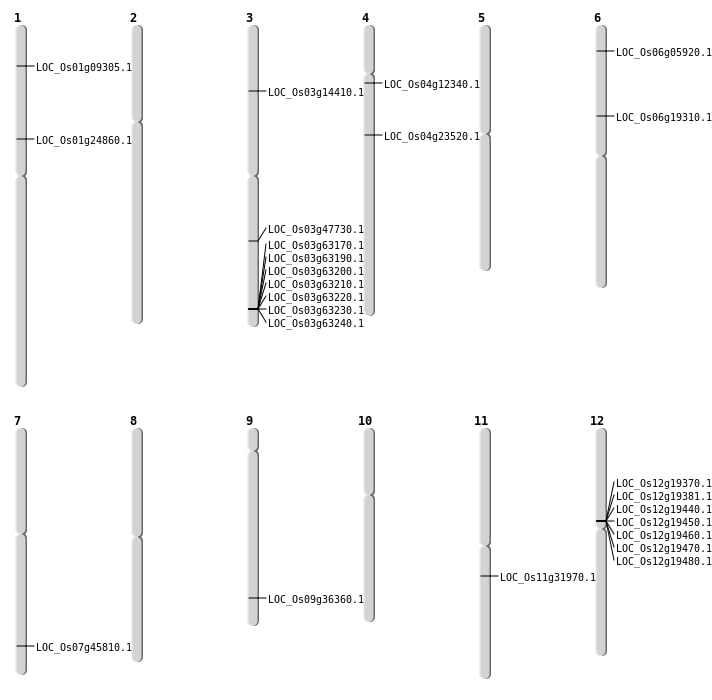
|
| Variant Information |
|---|
Single base substitutions: 45
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14002596 | C-G | HET | STOP_GAINED | LOC_Os01g24860 |
| SBS | Chr1 | 29237735 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 32147874 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 37483809 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 4732351 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g09305 |
| SBS | Chr1 | 49483 | C-T | HET | INTRON | |
| SBS | Chr10 | 10075169 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 3955438 | G-A | HET | INTRON | |
| SBS | Chr11 | 18316864 | A-G | HET | INTRON | |
| SBS | Chr11 | 4475186 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 1700187 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr12 | 25866858 | T-G | HET | INTERGENIC | |
| SBS | Chr12 | 27383223 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 10689479 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 18742333 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 19845978 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 27050798 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 2926696 | A-G | HET | INTRON | |
| SBS | Chr3 | 35894609 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 7837705 | G-T | HET | STOP_GAINED | LOC_Os03g14410 |
| SBS | Chr3 | 7837706 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g14410 |
| SBS | Chr4 | 11752493 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 13452447 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g23520 |
| SBS | Chr4 | 13548383 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 15965200 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 29884868 | A-G | HOMO | START_GAINED | |
| SBS | Chr4 | 4967720 | T-C | HET | INTRON | |
| SBS | Chr4 | 6809689 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g12340 |
| SBS | Chr5 | 28299404 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr5 | 28299405 | G-T | HET | UTR_3_PRIME | |
| SBS | Chr5 | 28299406 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr6 | 21756477 | T-C | HOMO | INTRON | |
| SBS | Chr6 | 2710292 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g05920 |
| SBS | Chr7 | 14098972 | C-T | HET | INTRON | |
| SBS | Chr7 | 21272723 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 25913627 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 26047518 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 6887092 | C-A | HET | INTRON | |
| SBS | Chr7 | 6887093 | A-G | HET | INTRON | |
| SBS | Chr8 | 12968546 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 26411402 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 6020906 | T-A | HET | SPLICE_SITE_REGION | |
| SBS | Chr8 | 7895571 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 16196216 | G-T | HET | INTRON | |
| SBS | Chr9 | 1711539 | G-C | HET | SYNONYMOUS_CODING |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 1104389 | 1104390 | 1 | |
| Deletion | Chr5 | 1440475 | 1440477 | 2 | |
| Deletion | Chr8 | 2724643 | 2724644 | 1 | |
| Deletion | Chr8 | 5824609 | 5824611 | 2 | |
| Deletion | Chr7 | 6344668 | 6344673 | 5 | |
| Deletion | Chr2 | 10522458 | 10522463 | 5 | |
| Deletion | Chr4 | 10630833 | 10630834 | 1 | |
| Deletion | Chr12 | 11252001 | 11272000 | 20000 | 2 |
| Deletion | Chr12 | 11290001 | 11331000 | 41000 | 5 |
| Deletion | Chr6 | 12208940 | 12208941 | 1 | |
| Deletion | Chr8 | 15396706 | 15396707 | 1 | |
| Deletion | Chr2 | 17034940 | 17035035 | 95 | |
| Deletion | Chr11 | 18799919 | 18799921 | 2 | LOC_Os11g31970 |
| Deletion | Chr9 | 18823226 | 18823227 | 1 | |
| Deletion | Chr8 | 19271780 | 19271781 | 1 | |
| Deletion | Chr5 | 21261806 | 21261807 | 1 | |
| Deletion | Chr11 | 22074866 | 22074871 | 5 | |
| Deletion | Chr10 | 22292810 | 22292814 | 4 | |
| Deletion | Chr12 | 22999369 | 22999374 | 5 | |
| Deletion | Chr4 | 30825491 | 30825578 | 87 | |
| Deletion | Chr3 | 35706001 | 35723000 | 17000 | 4 |
| Deletion | Chr3 | 35726001 | 35733000 | 7000 | 3 |
Insertions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 16199954 | 16199954 | 1 | |
| Insertion | Chr12 | 1803275 | 1803275 | 1 | |
| Insertion | Chr12 | 19681170 | 19681170 | 1 | |
| Insertion | Chr12 | 23863284 | 23863289 | 6 | |
| Insertion | Chr12 | 8491249 | 8491249 | 1 | |
| Insertion | Chr4 | 10817734 | 10817734 | 1 | |
| Insertion | Chr5 | 19153826 | 19153850 | 25 | |
| Insertion | Chr5 | 26747253 | 26747253 | 1 | |
| Insertion | Chr6 | 10982955 | 10982955 | 1 | LOC_Os06g19310 |
| Insertion | Chr6 | 29215735 | 29215735 | 1 | |
| Insertion | Chr7 | 27342456 | 27342458 | 3 | LOC_Os07g45810 |
| Insertion | Chr7 | 6535574 | 6535574 | 1 | |
| Insertion | Chr9 | 6149586 | 6149604 | 19 | |
| Insertion | Chr9 | 7293226 | 7293301 | 76 |
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 20615608 | 20983158 | |
| Inversion | Chr9 | 20985741 | 21027042 | LOC_Os09g36360 |
| Inversion | Chr8 | 21065454 | 21065870 | |
| Inversion | Chr3 | 27051204 | 27295370 | LOC_Os03g47730 |
| Inversion | Chr3 | 27051214 | 27295379 | LOC_Os03g47730 |
| Inversion | Chr1 | 31606402 | 31606567 | |
| Inversion | Chr3 | 35265592 | 35706611 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 20983160 | Chr4 | 29253461 | |
| Translocation | Chr9 | 20985731 | Chr4 | 29253460 | LOC_Os09g36360 |
