Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4607-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4607-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 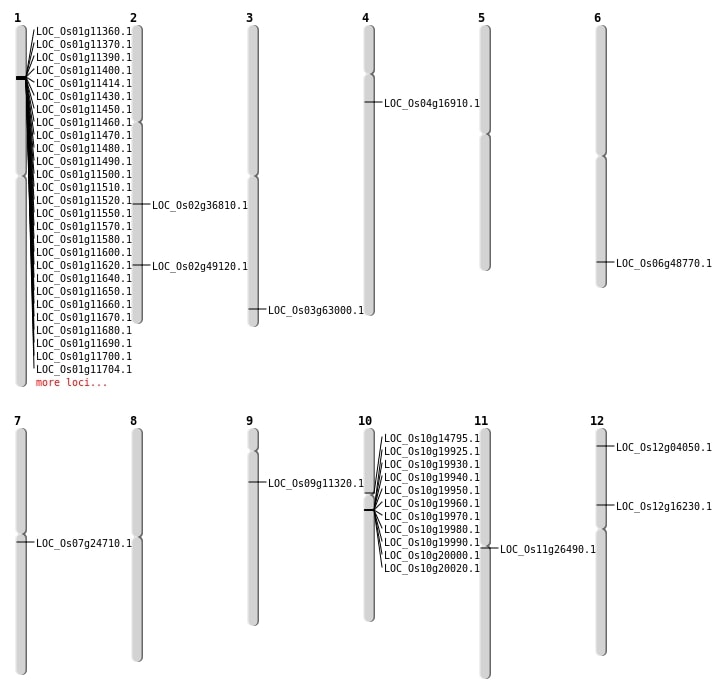
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10587294 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 5602865 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 5958846 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 10043080 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g20020 |
| SBS | Chr10 | 1820324 | A-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 7750456 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g14795 |
| SBS | Chr10 | 9029743 | T-G | HET | INTERGENIC | |
| SBS | Chr11 | 15186049 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g26490 |
| SBS | Chr11 | 17833607 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 1702520 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g04050 |
| SBS | Chr12 | 9285107 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g16230 |
| SBS | Chr2 | 11838832 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 30036708 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g49120 |
| SBS | Chr3 | 15848132 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 15848134 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 23113720 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 25382334 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 6036445 | T-C | HET | INTRON | |
| SBS | Chr5 | 1104122 | A-C | HOMO | INTRON | |
| SBS | Chr5 | 12834837 | T-C | HOMO | INTRON | |
| SBS | Chr5 | 18661103 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 22209474 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 11859588 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 16291221 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 24647684 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 29516905 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g48770 |
| SBS | Chr6 | 29516906 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g48770 |
| SBS | Chr7 | 14058264 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g24710 |
| SBS | Chr7 | 16693543 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 25385984 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 27246669 | G-A | HET | SPLICE_SITE_REGION | |
| SBS | Chr8 | 25630710 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 1232178 | C-A | HET | INTRON | |
| SBS | Chr9 | 13090259 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 13093846 | G-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 13093848 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 3709162 | T-G | HET | INTERGENIC |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 4654509 | 4654510 | 1 | |
| Deletion | Chr1 | 6103001 | 6401000 | 298000 | 42 |
| Deletion | Chr9 | 6285092 | 6285098 | 6 | LOC_Os09g11320 |
| Deletion | Chr8 | 6963019 | 6963020 | 1 | |
| Deletion | Chr9 | 8464435 | 8464436 | 1 | |
| Deletion | Chr6 | 9304704 | 9304708 | 4 | |
| Deletion | Chr5 | 9456893 | 9456898 | 5 | |
| Deletion | Chr10 | 9991001 | 10035000 | 44000 | 9 |
| Deletion | Chr5 | 10498656 | 10498677 | 21 | |
| Deletion | Chr2 | 11150894 | 11150899 | 5 | |
| Deletion | Chr11 | 12121961 | 12121962 | 1 | |
| Deletion | Chr2 | 16459247 | 16459314 | 67 | |
| Deletion | Chr3 | 16752683 | 16752686 | 3 | |
| Deletion | Chr8 | 17432896 | 17432897 | 1 | |
| Deletion | Chr7 | 21549231 | 21549233 | 2 | |
| Deletion | Chr2 | 22204172 | 22204176 | 4 | LOC_Os02g36810 |
| Deletion | Chr6 | 24481278 | 24481279 | 1 | |
| Deletion | Chr8 | 25298963 | 25298964 | 1 | |
| Deletion | Chr11 | 25740024 | 25740027 | 3 | |
| Deletion | Chr4 | 33575297 | 33575309 | 12 | |
| Deletion | Chr3 | 35622536 | 35622539 | 3 | LOC_Os03g63000 |
| Deletion | Chr1 | 41251001 | 41272000 | 21000 | LOC_Os01g71300 |
Insertions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 5026126 | 5026127 | 2 | |
| Insertion | Chr10 | 8574604 | 8574604 | 1 | |
| Insertion | Chr11 | 27727426 | 27727503 | 78 | |
| Insertion | Chr11 | 9987092 | 9987139 | 48 | |
| Insertion | Chr12 | 11689896 | 11689896 | 1 | |
| Insertion | Chr12 | 16228871 | 16228871 | 1 | |
| Insertion | Chr3 | 16385110 | 16385111 | 2 | |
| Insertion | Chr4 | 14963109 | 14963109 | 1 | |
| Insertion | Chr4 | 9253174 | 9253174 | 1 | LOC_Os04g16910 |
| Insertion | Chr7 | 28867623 | 28867623 | 1 | |
| Insertion | Chr8 | 12438243 | 12438243 | 1 | |
| Insertion | Chr8 | 17372438 | 17372439 | 2 | |
| Insertion | Chr8 | 28032789 | 28032789 | 1 | |
| Insertion | Chr9 | 14464181 | 14464181 | 1 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 10218202 | 10532432 | LOC_Os01g18240 |
| Inversion | Chr1 | 10218211 | 10532438 | LOC_Os01g18240 |
| Inversion | Chr12 | 24512866 | 24860885 | |
| Inversion | Chr1 | 24760063 | 24760234 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 20216144 | Chr1 | 42529151 | LOC_Os01g36420 |
