Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN4872-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN4872-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 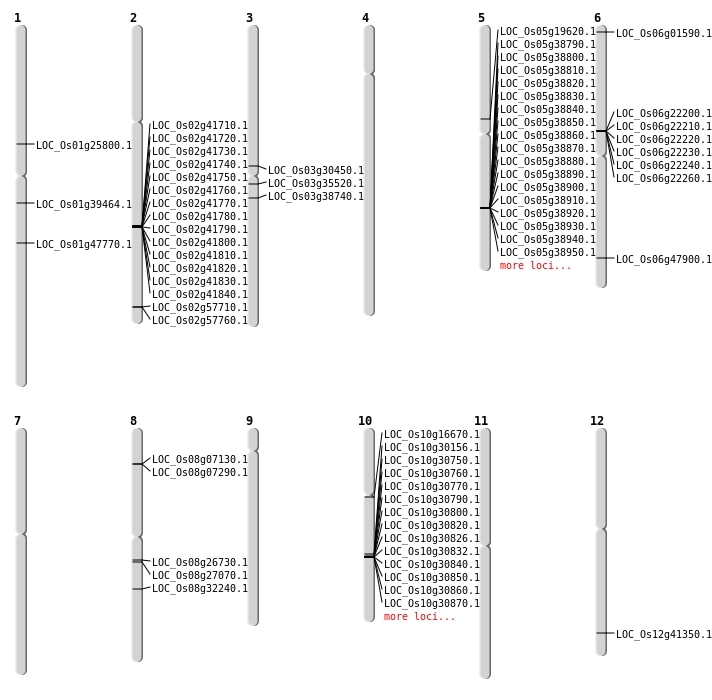
|
| Variant Information |
|---|
Single base substitutions: 50
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14603420 | G-C | HOMO | ||
| SBS | Chr1 | 14611368 | C-T | HOMO | LOC_Os01g25800 | |
| SBS | Chr1 | 14611372 | G-A | HOMO | LOC_Os01g25800 | |
| SBS | Chr1 | 21120832 | G-T | HET | ||
| SBS | Chr1 | 24191400 | C-A | HET | ||
| SBS | Chr1 | 27337303 | G-T | HET | LOC_Os01g47770 | |
| SBS | Chr1 | 28988811 | G-A | HET | ||
| SBS | Chr1 | 31195467 | C-T | HET | ||
| SBS | Chr1 | 40074637 | A-G | HET | ||
| SBS | Chr1 | 4757561 | G-A | HET | ||
| SBS | Chr10 | 6606342 | C-A | HET | ||
| SBS | Chr10 | 7701842 | A-T | HET | ||
| SBS | Chr10 | 8313959 | C-T | HET | LOC_Os10g16670 | |
| SBS | Chr11 | 17721043 | A-T | HOMO | ||
| SBS | Chr11 | 27678413 | G-A | HET | ||
| SBS | Chr12 | 13692477 | T-G | HET | ||
| SBS | Chr12 | 1912059 | G-A | HOMO | ||
| SBS | Chr12 | 22442640 | C-T | HET | ||
| SBS | Chr12 | 25718277 | C-A | HOMO | ||
| SBS | Chr2 | 18688409 | G-A | HET | ||
| SBS | Chr2 | 35373384 | A-G | HET | LOC_Os02g57760 | |
| SBS | Chr2 | 7348073 | G-A | HET | ||
| SBS | Chr3 | 19191737 | G-A | HET | ||
| SBS | Chr3 | 19686782 | A-T | HET | LOC_Os03g35520 | |
| SBS | Chr3 | 6106320 | A-C | HET | ||
| SBS | Chr4 | 11984242 | G-T | HET | ||
| SBS | Chr4 | 13196031 | G-A | HET | ||
| SBS | Chr4 | 21828019 | G-A | HET | ||
| SBS | Chr4 | 23205530 | T-A | HOMO | ||
| SBS | Chr5 | 11434206 | T-C | HET | LOC_Os05g19620 | |
| SBS | Chr5 | 6710307 | C-T | HET | ||
| SBS | Chr6 | 114527 | T-C | HET | ||
| SBS | Chr6 | 13673447 | C-A | HET | ||
| SBS | Chr6 | 21396519 | T-C | HET | ||
| SBS | Chr6 | 26519411 | C-T | HET | ||
| SBS | Chr6 | 27965814 | G-A | HET | ||
| SBS | Chr7 | 15128156 | A-T | HET | ||
| SBS | Chr7 | 17696653 | C-A | HOMO | ||
| SBS | Chr7 | 2372695 | A-G | HET | ||
| SBS | Chr7 | 551222 | A-T | HET | ||
| SBS | Chr8 | 11309943 | T-C | HET | ||
| SBS | Chr8 | 14131266 | G-A | HET | ||
| SBS | Chr8 | 1454952 | G-T | HOMO | ||
| SBS | Chr8 | 16260544 | G-A | HET | LOC_Os08g26730 | |
| SBS | Chr8 | 19977055 | G-A | HET | LOC_Os08g32240 | |
| SBS | Chr8 | 4076654 | C-T | HET | LOC_Os08g07290 | |
| SBS | Chr8 | 4685227 | C-A | HET | ||
| SBS | Chr8 | 6072687 | A-C | HET | ||
| SBS | Chr9 | 13250478 | C-T | HET | ||
| SBS | Chr9 | 18829377 | G-T | HET |
Deletions: 70
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 253772 | 253772 | 1 | |
| Deletion | Chr6 | 348267 | 348422 | 156 | LOC_Os06g01590 |
| Deletion | Chr8 | 775299 | 775340 | 42 | |
| Deletion | Chr6 | 1029607 | 1029612 | 6 | |
| Deletion | Chr4 | 2034630 | 2034631 | 2 | |
| Deletion | Chr8 | 2103561 | 2103586 | 26 | |
| Deletion | Chr9 | 2370519 | 2370534 | 16 | |
| Deletion | Chr8 | 2831615 | 2831619 | 5 | |
| Deletion | Chr7 | 3630565 | 3630572 | 8 | |
| Deletion | Chr4 | 3700240 | 3700240 | 1 | |
| Deletion | Chr8 | 3995432 | 3995482 | 51 | LOC_Os08g07130 |
| Deletion | Chr11 | 5200179 | 5200203 | 25 | |
| Deletion | Chr2 | 6294796 | 6294796 | 1 | |
| Deletion | Chr11 | 6320325 | 6320487 | 163 | |
| Deletion | Chr5 | 6434802 | 6434812 | 11 | |
| Deletion | Chr5 | 6765913 | 6765918 | 6 | |
| Deletion | Chr11 | 7812582 | 7812582 | 1 | |
| Deletion | Chr8 | 8142700 | 8142702 | 3 | |
| Deletion | Chr10 | 8557724 | 8557727 | 4 | |
| Deletion | Chr10 | 8920462 | 8920470 | 9 | |
| Deletion | Chr6 | 9711298 | 9711301 | 4 | |
| Deletion | Chr6 | 10042521 | 10042534 | 14 | |
| Deletion | Chr6 | 12879001 | 12921000 | 42000 | 6 |
| Deletion | Chr4 | 13072467 | 13072468 | 2 | |
| Deletion | Chr9 | 13941412 | 13941422 | 11 | |
| Deletion | Chr3 | 14716160 | 14716160 | 1 | |
| Deletion | Chr2 | 14856111 | 14856111 | 1 | |
| Deletion | Chr4 | 15135652 | 15135869 | 218 | |
| Deletion | Chr10 | 16015001 | 16151000 | 136000 | 18 |
| Deletion | Chr8 | 16458767 | 16458770 | 4 | |
| Deletion | Chr8 | 16538787 | 16538788 | 2 | LOC_Os08g27070 |
| Deletion | Chr2 | 16950516 | 16950516 | 1 | |
| Deletion | Chr3 | 17364983 | 17364985 | 3 | LOC_Os03g30450 |
| Deletion | Chr11 | 17728297 | 17728329 | 33 | |
| Deletion | Chr2 | 17832932 | 17832932 | 1 | |
| Deletion | Chr4 | 18347255 | 18347255 | 1 | |
| Deletion | Chr1 | 18594891 | 18594892 | 2 | |
| Deletion | Chr6 | 19551912 | 19551919 | 8 | |
| Deletion | Chr4 | 19604078 | 19604086 | 9 | |
| Deletion | Chr6 | 19712055 | 19712062 | 8 | |
| Deletion | Chr2 | 20016367 | 20016694 | 328 | |
| Deletion | Chr6 | 20136156 | 20136158 | 3 | |
| Deletion | Chr7 | 20725571 | 20725571 | 1 | |
| Deletion | Chr7 | 20777353 | 20777356 | 4 | |
| Deletion | Chr3 | 20803562 | 20803563 | 2 | |
| Deletion | Chr3 | 20886990 | 20886992 | 3 | |
| Deletion | Chr3 | 21502197 | 21502216 | 20 | LOC_Os03g38740 |
| Deletion | Chr1 | 22228238 | 22228276 | 39 | LOC_Os01g39464 |
| Deletion | Chr5 | 22508052 | 22508053 | 2 | |
| Deletion | Chr11 | 22751784 | 22751790 | 7 | |
| Deletion | Chr5 | 22753001 | 22883000 | 130000 | 22 |
| Deletion | Chr3 | 23129324 | 23129343 | 20 | |
| Deletion | Chr2 | 25045001 | 25155000 | 110000 | 14 |
| Deletion | Chr8 | 25552116 | 25552143 | 28 | |
| Deletion | Chr12 | 25627668 | 25627676 | 9 | LOC_Os12g41350 |
| Deletion | Chr7 | 25628745 | 25628752 | 8 | |
| Deletion | Chr5 | 25732012 | 25732012 | 1 | |
| Deletion | Chr12 | 26187457 | 26187488 | 32 | |
| Deletion | Chr2 | 27289974 | 27290017 | 44 | |
| Deletion | Chr8 | 27441796 | 27441798 | 3 | |
| Deletion | Chr11 | 27620747 | 27620845 | 99 | |
| Deletion | Chr6 | 28977950 | 28977950 | 1 | LOC_Os06g47900 |
| Deletion | Chr7 | 29320877 | 29320877 | 1 | |
| Deletion | Chr6 | 29337801 | 29337852 | 52 | |
| Deletion | Chr4 | 30820448 | 30820452 | 5 | |
| Deletion | Chr2 | 31226408 | 31226408 | 1 | |
| Deletion | Chr2 | 31619160 | 31619172 | 13 | |
| Deletion | Chr3 | 33496354 | 33496355 | 2 | |
| Deletion | Chr2 | 35345387 | 35345400 | 14 | LOC_Os02g57710 |
| Deletion | Chr3 | 36224916 | 36224917 | 2 |
Insertions: 34
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 1700804 | 1700931 | |
| Inversion | Chr12 | 13122681 | 13789299 | |
| Inversion | Chr12 | 13122689 | 13789300 | |
| Inversion | Chr10 | 15678098 | 15998869 | |
| Inversion | Chr10 | 15680265 | 16150490 | LOC_Os10g30156 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 14735412 | Chr7 | 15984557 | |
| Translocation | Chr11 | 14736087 | Chr7 | 15984578 |
