Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN490-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN490-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 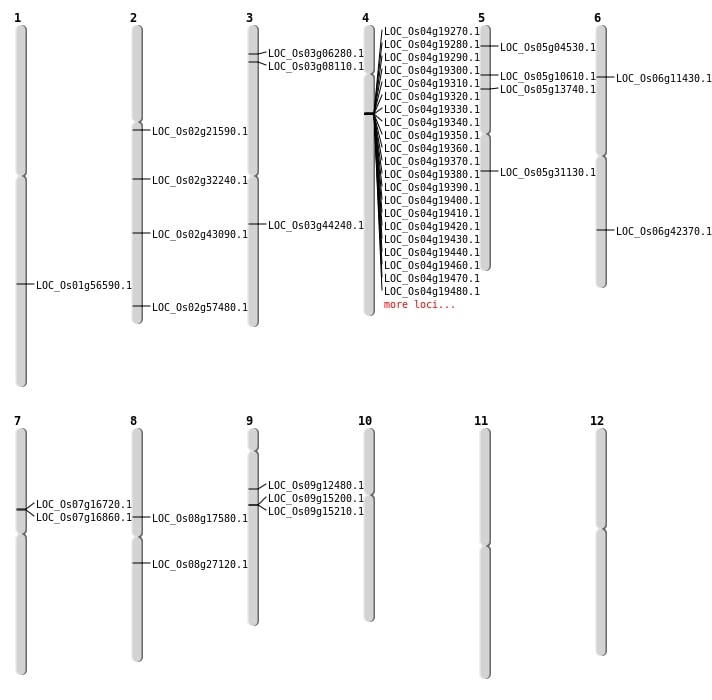
|
| Variant Information |
|---|
Single base substitutions: 25
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16141487 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 16141488 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 31942738 | T-G | HET | INTERGENIC | |
| SBS | Chr10 | 13108715 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 14914272 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 19082288 | A-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 9292667 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 3148820 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g06280 |
| SBS | Chr3 | 32482479 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 19640934 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 16878175 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 18094805 | G-A | HET | INTRON | |
| SBS | Chr5 | 18094806 | G-A | HET | INTRON | |
| SBS | Chr5 | 19786993 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 20943766 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 239217 | G-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 26899096 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 16704622 | A-G | HET | INTRON | |
| SBS | Chr6 | 5495712 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 10499034 | G-C | HET | INTERGENIC | |
| SBS | Chr7 | 17358016 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 9786289 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 13964633 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 25643283 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 11165165 | G-A | HET | SYNONYMOUS_CODING |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 783385 | 783392 | 7 | |
| Deletion | Chr5 | 2102781 | 2102794 | 13 | LOC_Os05g04530 |
| Deletion | Chr4 | 5023424 | 5023457 | 33 | |
| Deletion | Chr5 | 5782372 | 5782374 | 2 | LOC_Os05g10610 |
| Deletion | Chr9 | 6829524 | 6829528 | 4 | |
| Deletion | Chr5 | 7613987 | 7613993 | 6 | LOC_Os05g13740 |
| Deletion | Chr7 | 7924013 | 7924034 | 21 | |
| Deletion | Chr8 | 8912810 | 8912819 | 9 | |
| Deletion | Chr9 | 9228853 | 9235209 | 6356 | 2 |
| Deletion | Chr4 | 10707001 | 10890000 | 183000 | 29 |
| Deletion | Chr2 | 12824651 | 12824656 | 5 | LOC_Os02g21590 |
| Deletion | Chr5 | 13804073 | 13804074 | 1 | |
| Deletion | Chr5 | 15527185 | 15527186 | 1 | |
| Deletion | Chr8 | 16572355 | 16572357 | 2 | LOC_Os08g27120 |
| Deletion | Chr5 | 18099258 | 18099266 | 8 | LOC_Os05g31130 |
| Deletion | Chr8 | 18612794 | 18612796 | 2 | |
| Deletion | Chr2 | 19041253 | 19041254 | 1 | LOC_Os02g32240 |
| Deletion | Chr2 | 19234683 | 19234731 | 48 | |
| Deletion | Chr9 | 19243394 | 19243404 | 10 | |
| Deletion | Chr4 | 19498802 | 19498807 | 5 | LOC_Os04g32490 |
| Deletion | Chr4 | 21261300 | 21261301 | 1 | |
| Deletion | Chr3 | 24861723 | 24861731 | 8 | LOC_Os03g44240 |
| Deletion | Chr6 | 25468156 | 25468162 | 6 | LOC_Os06g42370 |
| Deletion | Chr2 | 25952255 | 25952257 | 2 | LOC_Os02g43090 |
| Deletion | Chr5 | 26899000 | 26899006 | 6 | |
| Deletion | Chr1 | 28561197 | 28561199 | 2 | |
| Deletion | Chr1 | 28633686 | 28633687 | 1 | |
| Deletion | Chr4 | 28833472 | 28833477 | 5 | |
| Deletion | Chr2 | 35223680 | 35223690 | 10 | LOC_Os02g57480 |
Insertions: 7
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 3142676 | 3142677 | 2 | |
| Insertion | Chr11 | 1897182 | 1897183 | 2 | |
| Insertion | Chr2 | 15314956 | 15314958 | 3 | |
| Insertion | Chr4 | 988630 | 988630 | 1 | |
| Insertion | Chr6 | 6032714 | 6032714 | 1 | LOC_Os06g11430 |
| Insertion | Chr7 | 11740182 | 11740183 | 2 | |
| Insertion | Chr7 | 23386232 | 23386233 | 2 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 7135469 | 8290314 | LOC_Os09g12480 |
| Inversion | Chr7 | 9814302 | 9913521 | 2 |
| Inversion | Chr7 | 9836063 | 9913526 | LOC_Os07g16860 |
| Inversion | Chr5 | 16618582 | 16912988 | |
| Inversion | Chr1 | 20603224 | 23080540 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 4141741 | Chr2 | 17320616 | LOC_Os03g08110 |
| Translocation | Chr9 | 9234527 | Chr1 | 32632617 | 2 |
| Translocation | Chr9 | 9235200 | Chr1 | 32632613 | LOC_Os01g56590 |
| Translocation | Chr7 | 9836050 | Chr4 | 13688950 | LOC_Os04g23930 |
| Translocation | Chr8 | 10754473 | Chr6 | 19260583 | LOC_Os08g17580 |
| Translocation | Chr4 | 13688963 | Chr3 | 928732 | LOC_Os04g23930 |
| Translocation | Chr4 | 13690611 | Chr3 | 928698 | LOC_Os04g23940 |
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr9 | 8714036 | 9363157 | 649121 |
