Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN495-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN495-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 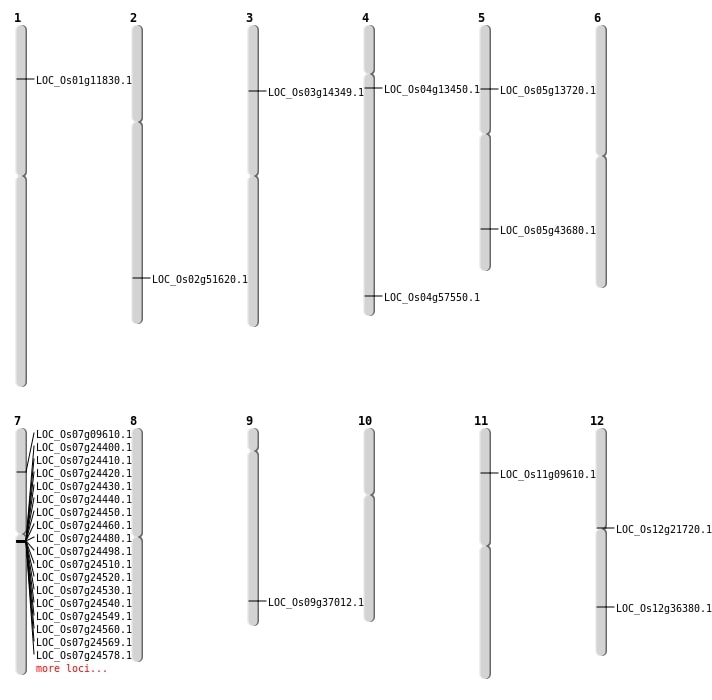
|
| Variant Information |
|---|
Single base substitutions: 29
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10837317 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 15195189 | C-A | HET | INTRON | |
| SBS | Chr1 | 19536118 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 32981318 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 33408685 | G-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 718524 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 13619028 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 21054395 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 3574373 | G-C | HET | INTERGENIC | |
| SBS | Chr10 | 6194607 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 15044954 | C-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 5138956 | A-T | HOMO | STOP_GAINED | LOC_Os11g09610 |
| SBS | Chr12 | 21426081 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 22272343 | T-G | HOMO | STOP_GAINED | LOC_Os12g36380 |
| SBS | Chr2 | 21595622 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 25472977 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 27750667 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 30370446 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 17950884 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 15526245 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 16531840 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 34242041 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g57550 |
| SBS | Chr5 | 11233075 | T-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 499964 | C-T | HET | INTRON | |
| SBS | Chr8 | 15996787 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 20341374 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 21188220 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 7759038 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 9061689 | A-T | HET | INTERGENIC |
Deletions: 35
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 659277 | 659294 | 17 | |
| Deletion | Chr11 | 2855070 | 2855083 | 13 | |
| Deletion | Chr4 | 4031555 | 4031557 | 2 | |
| Deletion | Chr1 | 4304915 | 4304916 | 1 | |
| Deletion | Chr7 | 5104661 | 5104666 | 5 | LOC_Os07g09610 |
| Deletion | Chr6 | 5728126 | 5728130 | 4 | |
| Deletion | Chr1 | 6396415 | 6396425 | 10 | LOC_Os01g11830 |
| Deletion | Chr5 | 6776726 | 6776732 | 6 | |
| Deletion | Chr4 | 7488443 | 7488446 | 3 | LOC_Os04g13450 |
| Deletion | Chr5 | 7599634 | 7599642 | 8 | LOC_Os05g13720 |
| Deletion | Chr2 | 7679806 | 7679807 | 1 | |
| Deletion | Chr3 | 7805970 | 7805971 | 1 | LOC_Os03g14349 |
| Deletion | Chr10 | 8688636 | 8688650 | 14 | |
| Deletion | Chr2 | 11311985 | 11311986 | 1 | |
| Deletion | Chr12 | 12213251 | 12213257 | 6 | LOC_Os12g21720 |
| Deletion | Chr12 | 12695873 | 12695892 | 19 | |
| Deletion | Chr7 | 13889001 | 14016000 | 127000 | 22 |
| Deletion | Chr6 | 14825140 | 14825149 | 9 | |
| Deletion | Chr5 | 18979214 | 18979218 | 4 | |
| Deletion | Chr5 | 18979218 | 18979222 | 4 | |
| Deletion | Chr10 | 20264874 | 20264908 | 34 | |
| Deletion | Chr11 | 20465940 | 20465950 | 10 | |
| Deletion | Chr2 | 20647504 | 20647520 | 16 | |
| Deletion | Chr9 | 21178913 | 21178930 | 17 | |
| Deletion | Chr9 | 21347675 | 21347682 | 7 | LOC_Os09g37012 |
| Deletion | Chr8 | 22452168 | 22452170 | 2 | |
| Deletion | Chr8 | 22537092 | 22537111 | 19 | |
| Deletion | Chr12 | 23209988 | 23209996 | 8 | |
| Deletion | Chr5 | 25395997 | 25396005 | 8 | LOC_Os05g43680 |
| Deletion | Chr12 | 26157295 | 26157298 | 3 | |
| Deletion | Chr2 | 29211569 | 29212694 | 1125 | |
| Deletion | Chr2 | 29311572 | 29311573 | 1 | |
| Deletion | Chr4 | 30031736 | 30031740 | 4 | |
| Deletion | Chr2 | 31609640 | 31609641 | 1 | LOC_Os02g51620 |
| Deletion | Chr4 | 35433829 | 35433833 | 4 |
Insertions: 5
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 23471544 | 23861646 | LOC_Os06g39520 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 20684291 | Chr1 | 38624282 |
