Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN499-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN499-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 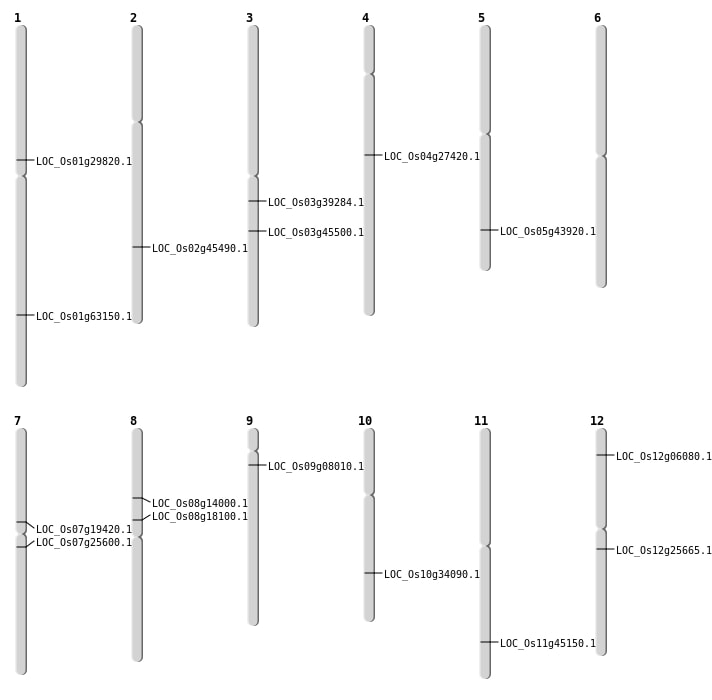
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 31405588 | A-T | HOMO | INTRON | |
| SBS | Chr1 | 32718221 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 36589028 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g63150 |
| SBS | Chr1 | 37789768 | A-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 12729315 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 17639610 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 19273482 | C-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 232458 | G-A | HET | INTRON | |
| SBS | Chr11 | 15644654 | G-A | HOMO | INTRON | |
| SBS | Chr12 | 9012103 | C-T | HET | INTRON | |
| SBS | Chr2 | 13309377 | G-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 13309378 | G-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 20212545 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 33092372 | A-T | HOMO | INTRON | |
| SBS | Chr3 | 1014561 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 13770164 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 18798440 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 21827852 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g39284 |
| SBS | Chr3 | 25678678 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g45500 |
| SBS | Chr3 | 581124 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 7127327 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 12547853 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 22559031 | C-T | HOMO | INTRON | |
| SBS | Chr4 | 6712162 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 13650792 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 24201689 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 13315232 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr6 | 24522433 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr6 | 29877413 | A-C | HOMO | INTRON | |
| SBS | Chr7 | 11489776 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g19420 |
| SBS | Chr7 | 14221243 | G-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 14677755 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g25600 |
| SBS | Chr7 | 18088502 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 18088504 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 21808139 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 11097979 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g18100 |
| SBS | Chr8 | 8396479 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g14000 |
| SBS | Chr8 | 8396480 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g14000 |
| SBS | Chr9 | 18045196 | T-C | HET | INTRON | |
| SBS | Chr9 | 22352886 | C-T | HET | INTRON | |
| SBS | Chr9 | 4125878 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os09g08010 |
| SBS | Chr9 | 4149028 | G-A | HOMO | INTERGENIC |
Deletions: 20
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 2258866 | 2258873 | 7 | |
| Deletion | Chr12 | 2836412 | 2836417 | 5 | LOC_Os12g06080 |
| Deletion | Chr2 | 5386328 | 5386338 | 10 | |
| Deletion | Chr12 | 6558243 | 6558251 | 8 | |
| Deletion | Chr7 | 13266444 | 13266448 | 4 | |
| Deletion | Chr2 | 13313190 | 13313195 | 5 | |
| Deletion | Chr12 | 14868300 | 14868304 | 4 | LOC_Os12g25665 |
| Deletion | Chr4 | 16211005 | 16211006 | 1 | LOC_Os04g27420 |
| Deletion | Chr1 | 16706655 | 16706673 | 18 | LOC_Os01g29820 |
| Deletion | Chr3 | 20949506 | 20949514 | 8 | |
| Deletion | Chr7 | 21808149 | 21808150 | 1 | |
| Deletion | Chr5 | 24202932 | 24202951 | 19 | |
| Deletion | Chr5 | 24958632 | 24958637 | 5 | |
| Deletion | Chr6 | 25258697 | 25258705 | 8 | |
| Deletion | Chr5 | 25558503 | 25558516 | 13 | LOC_Os05g43920 |
| Deletion | Chr8 | 27113579 | 27113591 | 12 | |
| Deletion | Chr11 | 27329765 | 27329766 | 1 | LOC_Os11g45150 |
| Deletion | Chr2 | 27670622 | 27670627 | 5 | LOC_Os02g45490 |
| Deletion | Chr3 | 29889732 | 29889755 | 23 | |
| Deletion | Chr2 | 30584679 | 30584684 | 5 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 1623145 | 1623146 | 2 | |
| Insertion | Chr11 | 28660277 | 28660278 | 2 | |
| Insertion | Chr6 | 3459409 | 3459409 | 1 | |
| Insertion | Chr9 | 13061771 | 13061772 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 34663594 | 35090864 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 18196670 | Chr9 | 3015086 | LOC_Os10g34090 |
