Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN509-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN509-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 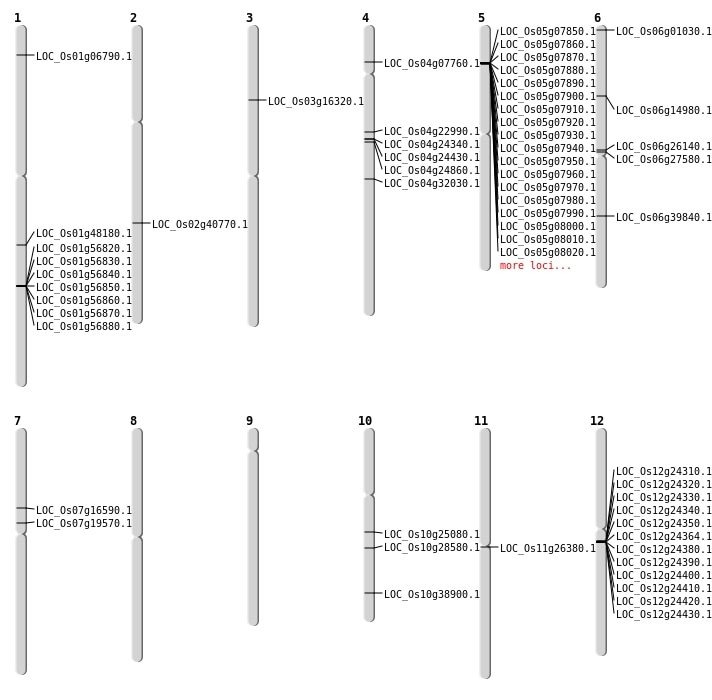
|
| Variant Information |
|---|
Single base substitutions: 45
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 27598377 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g48180 |
| SBS | Chr1 | 31922713 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 12912980 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g25080 |
| SBS | Chr10 | 1349709 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 12560644 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 15161256 | C-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 778173 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 19580079 | A-T | HET | INTRON | |
| SBS | Chr2 | 5761468 | G-C | HET | INTRON | |
| SBS | Chr2 | 9335635 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 18711105 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 21010215 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 22196267 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 23707411 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 30215260 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 7038444 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 7038445 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 13051799 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g22990 |
| SBS | Chr4 | 13246538 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 13279728 | A-T | HET | INTRON | |
| SBS | Chr4 | 14013054 | C-T | HET | STOP_GAINED | LOC_Os04g24430 |
| SBS | Chr4 | 14329900 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g24860 |
| SBS | Chr4 | 2101959 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 3192493 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 12122345 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 16222311 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 22920966 | A-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g39090 |
| SBS | Chr6 | 11323855 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 11718 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g01030 |
| SBS | Chr6 | 1242679 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 15295148 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g26140 |
| SBS | Chr6 | 21267118 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 6959951 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 7046653 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 8495210 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g14980 |
| SBS | Chr7 | 11598160 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g19570 |
| SBS | Chr7 | 14950484 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 23366294 | G-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 18808807 | C-T | HOMO | INTRON | |
| SBS | Chr8 | 21661599 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 7997814 | G-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 9191549 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 12286177 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 12286179 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 4936522 | G-T | HET | INTRON |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 663464 | 663466 | 2 | |
| Deletion | Chr1 | 1238423 | 1238424 | 1 | |
| Deletion | Chr6 | 1663588 | 1663610 | 22 | |
| Deletion | Chr9 | 3460792 | 3460802 | 10 | |
| Deletion | Chr5 | 4238001 | 4417000 | 179000 | 27 |
| Deletion | Chr10 | 6940381 | 6940384 | 3 | |
| Deletion | Chr7 | 8388856 | 8388858 | 2 | |
| Deletion | Chr3 | 9007439 | 9007445 | 6 | LOC_Os03g16320 |
| Deletion | Chr7 | 9731008 | 9731009 | 1 | LOC_Os07g16590 |
| Deletion | Chr4 | 10431609 | 10431621 | 12 | |
| Deletion | Chr5 | 11526661 | 11526670 | 9 | LOC_Os05g19750 |
| Deletion | Chr10 | 12101130 | 12101138 | 8 | |
| Deletion | Chr12 | 13870590 | 13946527 | 75937 | 12 |
| Deletion | Chr4 | 13950250 | 13950251 | 1 | LOC_Os04g24340 |
| Deletion | Chr10 | 14875277 | 14878003 | 2726 | LOC_Os10g28580 |
| Deletion | Chr11 | 15102067 | 15102073 | 6 | LOC_Os11g26380 |
| Deletion | Chr8 | 15504325 | 15504345 | 20 | |
| Deletion | Chr7 | 15952991 | 15952993 | 2 | |
| Deletion | Chr3 | 16081899 | 16082404 | 505 | |
| Deletion | Chr1 | 17347613 | 17347614 | 1 | |
| Deletion | Chr1 | 17347618 | 17347619 | 1 | |
| Deletion | Chr6 | 20856439 | 20856453 | 14 | |
| Deletion | Chr4 | 20933866 | 20933910 | 44 | |
| Deletion | Chr6 | 23663999 | 23664000 | 1 | LOC_Os06g39840 |
| Deletion | Chr11 | 24107461 | 24107473 | 12 | |
| Deletion | Chr2 | 24714010 | 24714011 | 1 | LOC_Os02g40770 |
| Deletion | Chr5 | 29117705 | 29117707 | 2 | |
| Deletion | Chr1 | 32794001 | 32854000 | 60000 | 7 |
| Deletion | Chr1 | 33171951 | 33171963 | 12 | |
| Deletion | Chr1 | 37978036 | 37978045 | 9 |
Insertions: 10
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 42459782 | 42459783 | 2 | |
| Insertion | Chr10 | 20730853 | 20730853 | 1 | LOC_Os10g38900 |
| Insertion | Chr11 | 23843690 | 23843690 | 1 | |
| Insertion | Chr2 | 35526121 | 35526122 | 2 | |
| Insertion | Chr3 | 1946352 | 1946352 | 1 | |
| Insertion | Chr4 | 4124944 | 4125002 | 59 | LOC_Os04g07760 |
| Insertion | Chr6 | 4464678 | 4464681 | 4 | |
| Insertion | Chr7 | 7529539 | 7529542 | 4 | |
| Insertion | Chr8 | 3347906 | 3347907 | 2 | |
| Insertion | Chr9 | 18262261 | 18262261 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 14014238 | 19000349 | |
| Inversion | Chr4 | 19000335 | 19191788 | LOC_Os04g32030 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 7565239 | Chr6 | 15609190 | LOC_Os06g27580 |
| Translocation | Chr9 | 12257861 | Chr1 | 3225642 | LOC_Os01g06790 |
| Translocation | Chr8 | 19838640 | Chr6 | 7023925 | |
| Translocation | Chr5 | 26902687 | Chr3 | 16081925 | LOC_Os05g46430 |
