Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN513-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN513-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 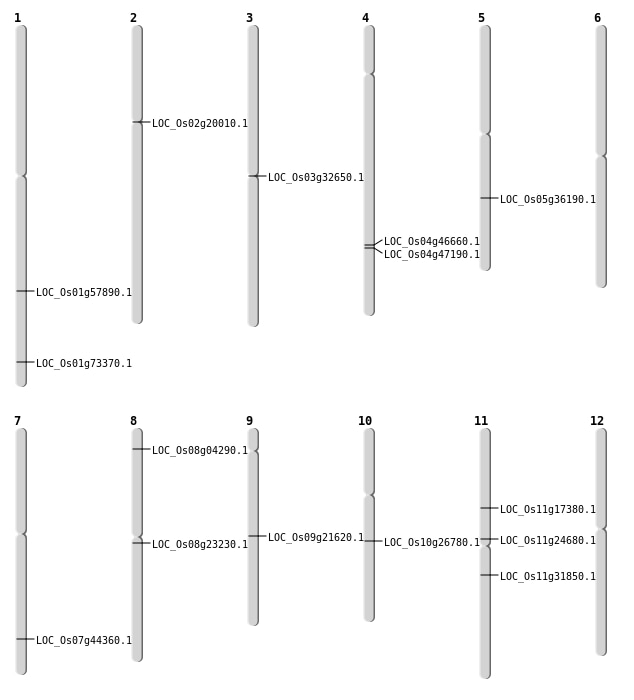
|
| Variant Information |
|---|
Single base substitutions: 27
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 19158079 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 35149900 | C-G | HOMO | INTRON | |
| SBS | Chr11 | 9691924 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g17380 |
| SBS | Chr12 | 5727367 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 11781793 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g20010 |
| SBS | Chr2 | 14454255 | A-C | HET | INTERGENIC | |
| SBS | Chr2 | 30187876 | C-A | HET | INTRON | |
| SBS | Chr3 | 18704438 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os03g32650 |
| SBS | Chr3 | 26639492 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 272783 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr3 | 29301829 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 15971012 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 27861517 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 23713355 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 28673894 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 18089296 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 18089297 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 28033817 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 6495093 | G-C | HET | UTR_3_PRIME | |
| SBS | Chr6 | 7712929 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 13578438 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 14026605 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g23230 |
| SBS | Chr8 | 1460882 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 17606950 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 17774715 | A-C | HET | INTRON | |
| SBS | Chr8 | 2507002 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 13089656 | G-C | HET | STOP_GAINED | LOC_Os09g21620 |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 2099284 | 2099298 | 14 | LOC_Os08g04290 |
| Deletion | Chr2 | 2140045 | 2140052 | 7 | |
| Deletion | Chr12 | 3058600 | 3058603 | 3 | |
| Deletion | Chr6 | 3328256 | 3328257 | 1 | |
| Deletion | Chr3 | 4421687 | 4421688 | 1 | |
| Deletion | Chr9 | 6354306 | 6354323 | 17 | |
| Deletion | Chr12 | 8402791 | 8402800 | 9 | |
| Deletion | Chr9 | 10940797 | 10940798 | 1 | |
| Deletion | Chr11 | 11380991 | 11380995 | 4 | |
| Deletion | Chr1 | 11851713 | 11851723 | 10 | |
| Deletion | Chr10 | 13108528 | 13108538 | 10 | |
| Deletion | Chr11 | 14073482 | 14073613 | 131 | LOC_Os11g24680 |
| Deletion | Chr12 | 15177444 | 15177467 | 23 | |
| Deletion | Chr7 | 16294132 | 16294137 | 5 | |
| Deletion | Chr11 | 18719074 | 18719082 | 8 | LOC_Os11g31850 |
| Deletion | Chr5 | 21463352 | 21463353 | 1 | LOC_Os05g36190 |
| Deletion | Chr6 | 21609034 | 21609035 | 1 | |
| Deletion | Chr4 | 23823145 | 23823150 | 5 | |
| Deletion | Chr6 | 24479081 | 24479095 | 14 | |
| Deletion | Chr7 | 26508513 | 26508529 | 16 | LOC_Os07g44360 |
| Deletion | Chr8 | 27328970 | 27328982 | 12 | |
| Deletion | Chr4 | 27662308 | 27662312 | 4 | LOC_Os04g46660 |
| Deletion | Chr4 | 28026682 | 28026686 | 4 | LOC_Os04g47190 |
| Deletion | Chr1 | 34999437 | 34999447 | 10 | |
| Deletion | Chr1 | 40533381 | 40533382 | 1 | |
| Deletion | Chr1 | 42527158 | 42527164 | 6 | LOC_Os01g73370 |
Insertions: 7
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 2756321 | 2756683 | |
| Inversion | Chr4 | 4168998 | 4169160 | |
| Inversion | Chr4 | 10843284 | 10843575 | |
| Inversion | Chr10 | 14011235 | 14011421 | LOC_Os10g26780 |
| Inversion | Chr4 | 21055885 | 21056053 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 4219528 | Chr1 | 33474600 | LOC_Os01g57890 |
| Translocation | Chr11 | 6352914 | Chr3 | 3398122 | |
| Translocation | Chr11 | 6353075 | Chr3 | 3397984 |
