Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN543-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN543-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 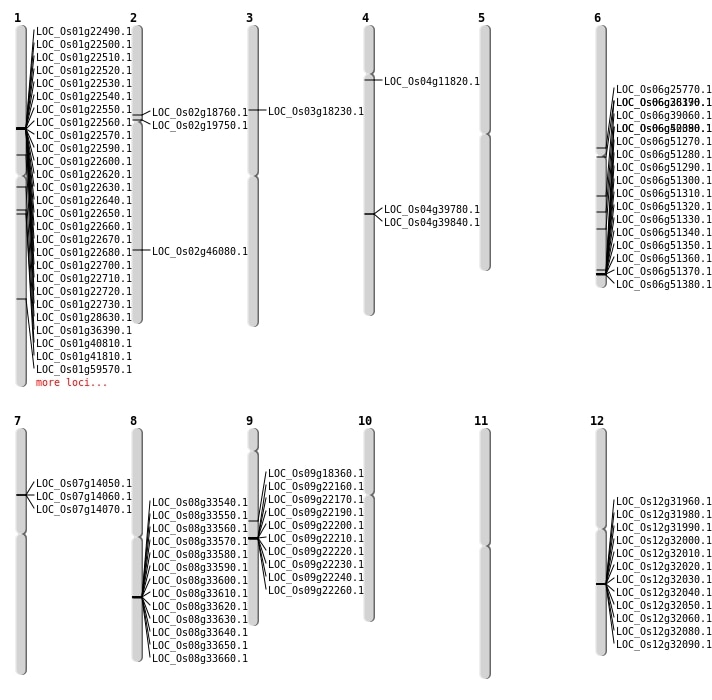
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 20194816 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g36390 |
| SBS | Chr1 | 22977865 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 23057760 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g40810 |
| SBS | Chr1 | 34410174 | A-G | HOMO | INTRON | |
| SBS | Chr1 | 37725889 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g64990 |
| SBS | Chr10 | 10211551 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 3835638 | T-C | HOMO | INTRON | |
| SBS | Chr11 | 14727703 | G-A | HOMO | INTRON | |
| SBS | Chr11 | 1530614 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 1530615 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 24115647 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 11638945 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 15108711 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 1866270 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 10061057 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 27431507 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 3032084 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 30990054 | C-A | HET | UTR_5_PRIME | |
| SBS | Chr3 | 4031023 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 14097914 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 6478341 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g11820 |
| SBS | Chr4 | 7247804 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 8338428 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 8683531 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 14050171 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 26204153 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 10178722 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 14434436 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 15054009 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g25770 |
| SBS | Chr6 | 19822057 | A-C | HET | INTERGENIC | |
| SBS | Chr6 | 23188725 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g39060 |
| SBS | Chr8 | 14060406 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 15305729 | A-G | HET | INTRON | |
| SBS | Chr8 | 20920353 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 23797208 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 11293830 | C-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 2126892 | A-C | HOMO | INTRON |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 1748114 | 1748125 | 11 | |
| Deletion | Chr6 | 2086377 | 2086388 | 11 | |
| Deletion | Chr3 | 2766967 | 2766976 | 9 | |
| Deletion | Chr12 | 3064927 | 3064928 | 1 | |
| Deletion | Chr6 | 5985021 | 5985023 | 2 | |
| Deletion | Chr7 | 8017003 | 8023711 | 6708 | 3 |
| Deletion | Chr9 | 11265774 | 11265793 | 19 | LOC_Os09g18360 |
| Deletion | Chr1 | 12644001 | 12780000 | 136000 | 22 |
| Deletion | Chr9 | 13413001 | 13468000 | 55000 | 9 |
| Deletion | Chr1 | 16015598 | 16020883 | 5285 | LOC_Os01g28630 |
| Deletion | Chr3 | 17806924 | 17806938 | 14 | |
| Deletion | Chr1 | 18140211 | 18140212 | 1 | |
| Deletion | Chr12 | 19262944 | 19362108 | 99164 | 12 |
| Deletion | Chr8 | 20937001 | 21024000 | 87000 | 13 |
| Deletion | Chr8 | 22820963 | 22820977 | 14 | |
| Deletion | Chr1 | 23203403 | 23203411 | 8 | |
| Deletion | Chr11 | 23480500 | 23480502 | 2 | |
| Deletion | Chr1 | 23670623 | 23675121 | 4498 | LOC_Os01g41810 |
| Deletion | Chr6 | 26562729 | 26562756 | 27 | |
| Deletion | Chr3 | 29666302 | 29666303 | 1 | |
| Deletion | Chr2 | 30713959 | 30713961 | 2 | |
| Deletion | Chr6 | 31031001 | 31121000 | 90000 | 12 |
| Deletion | Chr1 | 40201222 | 40201236 | 14 | |
| Deletion | Chr1 | 41947131 | 41947133 | 2 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 28081723 | 28081724 | 2 | LOC_Os02g46080 |
| Insertion | Chr3 | 10224923 | 10224924 | 2 | LOC_Os03g18230 |
| Insertion | Chr5 | 20720242 | 20720242 | 1 | |
| Insertion | Chr6 | 21171225 | 21171225 | 1 | LOC_Os06g36170 |
| Insertion | Chr6 | 6202864 | 6202865 | 2 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 19992228 | 21023893 | |
| Inversion | Chr8 | 20489729 | 20936931 | |
| Inversion | Chr6 | 20733539 | 25282498 | LOC_Os06g42090 |
| Inversion | Chr1 | 29814892 | 34448233 | LOC_Os01g59570 |
| Inversion | Chr6 | 30497346 | 31120902 | 2 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 8958194 | Chr1 | 23675098 | LOC_Os01g41810 |
| Translocation | Chr9 | 8958196 | Chr1 | 23672195 | |
| Translocation | Chr2 | 11552943 | Chr1 | 23672182 | LOC_Os02g19750 |
| Translocation | Chr2 | 11552950 | Chr1 | 23670626 | LOC_Os02g19750 |
| Translocation | Chr6 | 16153380 | Chr2 | 10936403 | 2 |
| Translocation | Chr11 | 19638385 | Chr4 | 23729597 | LOC_Os04g39840 |
| Translocation | Chr11 | 19638387 | Chr4 | 23700637 | LOC_Os04g39780 |
| Translocation | Chr12 | 25499049 | Chr1 | 41499146 |
