Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN553-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN553-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 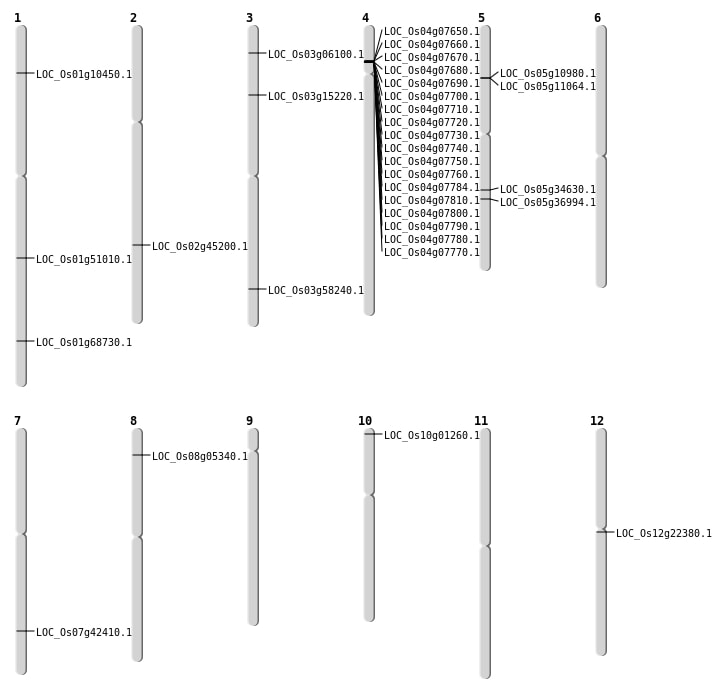
|
| Variant Information |
|---|
Single base substitutions: 25
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12858631 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 369768 | A-T | HET | INTRON | |
| SBS | Chr1 | 37128334 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 39486515 | T-C | HOMO | INTERGENIC | |
| SBS | Chr1 | 5511192 | A-T | HOMO | STOP_GAINED | LOC_Os01g10450 |
| SBS | Chr10 | 20820934 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 14708198 | A-G | HET | INTRON | |
| SBS | Chr11 | 18206368 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 23324428 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 25460512 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 19431769 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 13038481 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 3054865 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g06100 |
| SBS | Chr3 | 6635995 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 24670291 | G-A | HET | INTRON | |
| SBS | Chr4 | 24670293 | G-A | HET | INTRON | |
| SBS | Chr5 | 21151685 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 2125823 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 27074756 | T-A | HET | INTRON | |
| SBS | Chr7 | 29058470 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 24224132 | G-T | HOMO | INTRON | |
| SBS | Chr8 | 3396154 | T-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 6039042 | G-A | HOMO | UTR_3_PRIME | |
| SBS | Chr9 | 10832655 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 9150320 | A-G | HET | INTRON |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 1491105 | 1491106 | 1 | |
| Deletion | Chr11 | 2497856 | 2497857 | 1 | |
| Deletion | Chr8 | 2807051 | 2817513 | 10462 | LOC_Os08g05340 |
| Deletion | Chr8 | 3532913 | 3532914 | 1 | |
| Deletion | Chr4 | 4062001 | 4162000 | 100000 | 18 |
| Deletion | Chr11 | 4109056 | 4109064 | 8 | |
| Deletion | Chr4 | 5207752 | 5207753 | 1 | |
| Deletion | Chr5 | 7175068 | 7175070 | 2 | |
| Deletion | Chr3 | 8328547 | 8328548 | 1 | LOC_Os03g15220 |
| Deletion | Chr6 | 11213870 | 11213875 | 5 | |
| Deletion | Chr5 | 11701449 | 11701452 | 3 | |
| Deletion | Chr12 | 12641188 | 12641201 | 13 | LOC_Os12g22380 |
| Deletion | Chr8 | 13586300 | 13586316 | 16 | |
| Deletion | Chr5 | 14710107 | 14710143 | 36 | |
| Deletion | Chr10 | 18123041 | 18123042 | 1 | |
| Deletion | Chr8 | 18159784 | 18159791 | 7 | |
| Deletion | Chr10 | 19577414 | 19577417 | 3 | |
| Deletion | Chr5 | 20537863 | 20538867 | 1004 | LOC_Os05g34630 |
| Deletion | Chr5 | 23864715 | 23864719 | 4 | |
| Deletion | Chr7 | 25383170 | 25383171 | 1 | LOC_Os07g42410 |
| Deletion | Chr12 | 26244206 | 26244209 | 3 | |
| Deletion | Chr11 | 26756458 | 26756461 | 3 | |
| Deletion | Chr2 | 27436074 | 27436077 | 3 | LOC_Os02g45200 |
| Deletion | Chr4 | 27775412 | 27775428 | 16 | |
| Deletion | Chr1 | 29306907 | 29306913 | 6 | LOC_Os01g51010 |
| Deletion | Chr6 | 30375420 | 30375442 | 22 | |
| Deletion | Chr2 | 32225401 | 32225402 | 1 | |
| Deletion | Chr1 | 39923353 | 39923363 | 10 | LOC_Os01g68730 |
Insertions: 7
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 170813 | 936259 | LOC_Os10g01260 |
| Inversion | Chr5 | 6199776 | 21614264 | 2 |
| Inversion | Chr3 | 25907017 | 30952223 | |
| Inversion | Chr3 | 32172590 | 33183731 | LOC_Os03g58240 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 249606 | Chr5 | 6133758 | LOC_Os05g10980 |
| Translocation | Chr8 | 249873 | Chr5 | 6133728 | |
| Translocation | Chr12 | 4820674 | Chr9 | 18095065 | |
| Translocation | Chr5 | 20537879 | Chr1 | 29754991 | LOC_Os05g34630 |
| Translocation | Chr5 | 21422202 | Chr3 | 25354096 |
