Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN620-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN620-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 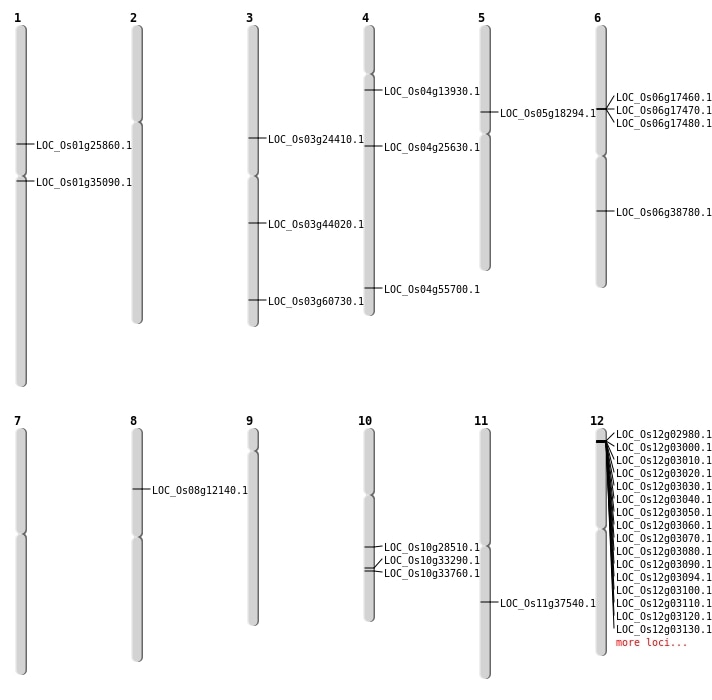
|
| Variant Information |
|---|
Single base substitutions: 47
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10636611 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 11359424 | G-T | HET | INTRON | |
| SBS | Chr1 | 18906859 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 41727195 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 1538854 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 17475354 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 19781574 | C-T | HOMO | INTRON | |
| SBS | Chr10 | 22547112 | C-T | HET | INTRON | |
| SBS | Chr10 | 8347260 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 8526813 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 14970991 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 15722388 | G-C | HET | INTRON | |
| SBS | Chr12 | 16695549 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 2383063 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 6948487 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 9036015 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g15850 |
| SBS | Chr12 | 9477672 | C-T | HET | INTRON | |
| SBS | Chr2 | 10115465 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 10115466 | C-G | HET | INTERGENIC | |
| SBS | Chr2 | 30637734 | A-T | HET | INTRON | |
| SBS | Chr3 | 24421306 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 24900208 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 13272462 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 13281011 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 14852637 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g25630 |
| SBS | Chr4 | 18805512 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 20291590 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 7782454 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g13930 |
| SBS | Chr4 | 8224758 | G-T | HET | INTRON | |
| SBS | Chr4 | 8224760 | A-T | HET | INTRON | |
| SBS | Chr5 | 13920627 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 1917247 | A-C | HET | INTERGENIC | |
| SBS | Chr5 | 1917248 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 13121216 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 24496115 | T-A | HET | INTRON | |
| SBS | Chr6 | 9815939 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 14515699 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 19414380 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 400937 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 11442254 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 26389584 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 7156399 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g12140 |
| SBS | Chr9 | 15503344 | T-A | HET | INTRON | |
| SBS | Chr9 | 15509014 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 20338033 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 2889146 | C-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 4812695 | G-T | HET | INTERGENIC |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 910959 | 912341 | 1382 | |
| Deletion | Chr12 | 1110001 | 1440000 | 330000 | 58 |
| Deletion | Chr6 | 2516831 | 2516832 | 1 | |
| Deletion | Chr10 | 4655410 | 4655412 | 2 | |
| Deletion | Chr10 | 7299202 | 7299209 | 7 | |
| Deletion | Chr11 | 7978609 | 7978611 | 2 | |
| Deletion | Chr3 | 8188400 | 8188401 | 1 | |
| Deletion | Chr12 | 8908849 | 8908875 | 26 | |
| Deletion | Chr6 | 10129001 | 10140000 | 11000 | 3 |
| Deletion | Chr3 | 11617468 | 11617469 | 1 | |
| Deletion | Chr5 | 12345929 | 12345952 | 23 | |
| Deletion | Chr9 | 13169499 | 13169500 | 1 | |
| Deletion | Chr3 | 13905928 | 13905929 | 1 | LOC_Os03g24410 |
| Deletion | Chr5 | 14448570 | 14448574 | 4 | |
| Deletion | Chr7 | 14515702 | 14515705 | 3 | |
| Deletion | Chr1 | 14667206 | 14667214 | 8 | LOC_Os01g25860 |
| Deletion | Chr11 | 14707582 | 14707583 | 1 | |
| Deletion | Chr10 | 17851231 | 17851241 | 10 | LOC_Os10g33760 |
| Deletion | Chr1 | 18456554 | 18456570 | 16 | |
| Deletion | Chr1 | 18918164 | 18918175 | 11 | |
| Deletion | Chr1 | 19431993 | 19431998 | 5 | LOC_Os01g35090 |
| Deletion | Chr3 | 19823641 | 19823645 | 4 | |
| Deletion | Chr10 | 20056173 | 20056174 | 1 | |
| Deletion | Chr6 | 23019948 | 23019949 | 1 | LOC_Os06g38780 |
| Deletion | Chr6 | 23298482 | 23298485 | 3 | |
| Deletion | Chr3 | 27219384 | 27219388 | 4 | |
| Deletion | Chr1 | 29739083 | 29739097 | 14 | |
| Deletion | Chr4 | 33152051 | 33152065 | 14 | LOC_Os04g55700 |
| Deletion | Chr3 | 34524547 | 34524552 | 5 | LOC_Os03g60730 |
Insertions: 8
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 2233149 | 2233152 | 4 | |
| Insertion | Chr11 | 22164614 | 22164614 | 1 | LOC_Os11g37540 |
| Insertion | Chr11 | 24415987 | 24415998 | 12 | |
| Insertion | Chr11 | 9613216 | 9613217 | 2 | |
| Insertion | Chr12 | 3632266 | 3632267 | 2 | |
| Insertion | Chr2 | 7763376 | 7763377 | 2 | |
| Insertion | Chr5 | 21765747 | 21765750 | 4 | |
| Insertion | Chr9 | 11029944 | 11029944 | 1 |
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 10546516 | 14986716 | LOC_Os05g18294 |
| Inversion | Chr5 | 10546529 | 14986724 | LOC_Os05g18294 |
| Inversion | Chr10 | 14834707 | 16345659 | LOC_Os10g28510 |
| Inversion | Chr10 | 14834720 | 16345693 | LOC_Os10g28510 |
| Inversion | Chr10 | 17185849 | 17487688 | LOC_Os10g33290 |
| Inversion | Chr3 | 24570278 | 24745233 | LOC_Os03g44020 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 3750283 | Chr5 | 912310 | |
| Translocation | Chr7 | 3750307 | Chr5 | 910968 |
