Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN633-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN633-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 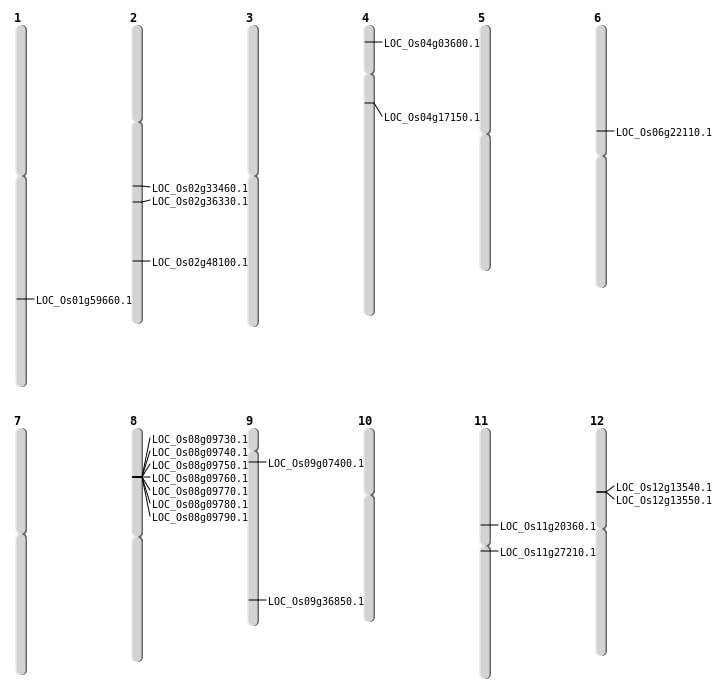
|
| Variant Information |
|---|
Single base substitutions: 27
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14207318 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 23412500 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 28046849 | G-A | HET | INTRON | |
| SBS | Chr11 | 13460725 | G-A | HET | UTR_3_PRIME | |
| SBS | Chr11 | 15663712 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g27210 |
| SBS | Chr11 | 3013525 | G-A | HOMO | INTRON | |
| SBS | Chr12 | 12446267 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 13319040 | G-C | HET | INTRON | |
| SBS | Chr12 | 14687019 | T-G | HET | INTERGENIC | |
| SBS | Chr12 | 22470436 | T-C | HET | INTERGENIC | |
| SBS | Chr12 | 24661229 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 8008121 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 8008123 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 21927858 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g36330 |
| SBS | Chr2 | 21927999 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g36330 |
| SBS | Chr2 | 31089883 | T-G | HOMO | INTRON | |
| SBS | Chr2 | 35404246 | T-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 25358315 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 30849820 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 14014457 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 12836975 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g22110 |
| SBS | Chr7 | 27218674 | T-C | HET | INTRON | |
| SBS | Chr7 | 27218822 | A-G | HOMO | INTRON | |
| SBS | Chr7 | 27218830 | C-T | HOMO | INTRON | |
| SBS | Chr8 | 1596374 | T-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 21832816 | G-C | HET | INTERGENIC | |
| SBS | Chr9 | 10782855 | A-G | HET | INTERGENIC |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 4593 | 4594 | 1 | |
| Deletion | Chr4 | 1573888 | 1573896 | 8 | LOC_Os04g03600 |
| Deletion | Chr9 | 2588589 | 2588591 | 2 | |
| Deletion | Chr3 | 3258012 | 3258021 | 9 | |
| Deletion | Chr9 | 3790679 | 3790688 | 9 | |
| Deletion | Chr3 | 5391103 | 5391114 | 11 | |
| Deletion | Chr8 | 5622001 | 5659000 | 37000 | 7 |
| Deletion | Chr9 | 7064485 | 7064488 | 3 | |
| Deletion | Chr12 | 7596605 | 7605534 | 8929 | 2 |
| Deletion | Chr10 | 8022132 | 8022138 | 6 | |
| Deletion | Chr4 | 9405935 | 9405946 | 11 | LOC_Os04g17150 |
| Deletion | Chr10 | 10149217 | 10149236 | 19 | |
| Deletion | Chr4 | 13192334 | 13192336 | 2 | |
| Deletion | Chr8 | 13566154 | 13566155 | 1 | |
| Deletion | Chr2 | 14406996 | 14407006 | 10 | |
| Deletion | Chr10 | 14469845 | 14469846 | 1 | |
| Deletion | Chr2 | 15159786 | 15159798 | 12 | |
| Deletion | Chr3 | 15178611 | 15178615 | 4 | |
| Deletion | Chr11 | 15567142 | 15567143 | 1 | |
| Deletion | Chr11 | 17660172 | 17660176 | 4 | |
| Deletion | Chr10 | 18372630 | 18372631 | 1 | |
| Deletion | Chr5 | 19078774 | 19078775 | 1 | |
| Deletion | Chr9 | 21252173 | 21252174 | 1 | LOC_Os09g36850 |
| Deletion | Chr12 | 26058787 | 26058789 | 2 | |
| Deletion | Chr2 | 29451361 | 29451363 | 2 | LOC_Os02g48100 |
| Deletion | Chr1 | 34511074 | 34511080 | 6 | LOC_Os01g59660 |
| Deletion | Chr1 | 35264855 | 35264856 | 1 | |
| Deletion | Chr1 | 40715222 | 40715223 | 1 | |
| Deletion | Chr1 | 41725456 | 41725458 | 2 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 24505789 | 24505790 | 2 | |
| Insertion | Chr6 | 20677302 | 20677303 | 2 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 3680902 | 3784051 | LOC_Os09g07400 |
| Inversion | Chr9 | 3680913 | 3784053 | LOC_Os09g07400 |
| Inversion | Chr11 | 10446630 | 11779307 | LOC_Os11g20360 |
| Inversion | Chr2 | 10674030 | 13599986 | |
| Inversion | Chr2 | 18843318 | 19895503 | LOC_Os02g33460 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 11739316 | Chr2 | 18843349 | |
| Translocation | Chr10 | 11752347 | Chr2 | 19895491 | LOC_Os02g33460 |
| Translocation | Chr5 | 14705825 | Chr2 | 12182619 |
