Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN661-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN661-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 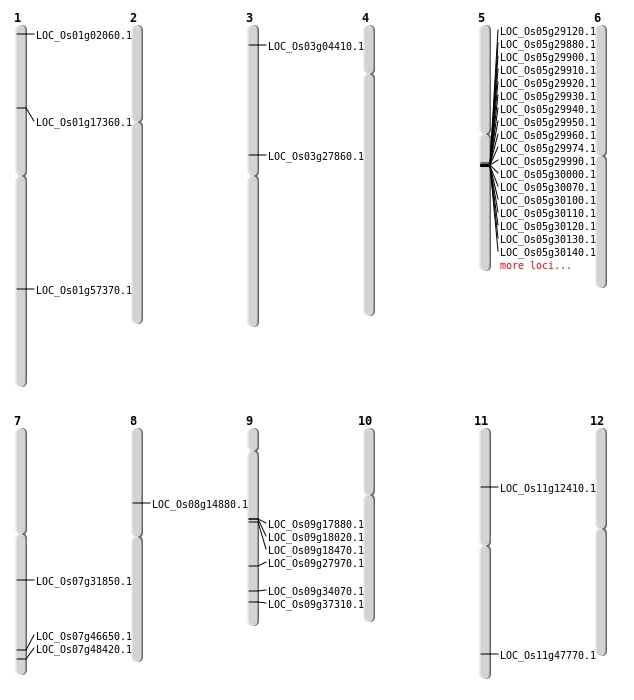
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 29659695 | C-G | HET | INTERGENIC | |
| SBS | Chr1 | 33225716 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 39881940 | A-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 39890875 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 249258 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 4490047 | G-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 22058255 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 10521026 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 14668210 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 15943162 | T-G | HET | INTERGENIC | |
| SBS | Chr12 | 17582163 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 20141285 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 25778826 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 9931646 | A-C | HET | INTERGENIC | |
| SBS | Chr2 | 11392242 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 20729150 | T-C | HET | INTRON | |
| SBS | Chr2 | 24752663 | C-A | HOMO | INTRON | |
| SBS | Chr2 | 6475396 | C-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 10703114 | G-T | HOMO | INTRON | |
| SBS | Chr3 | 1782730 | A-T | HET | INTERGENIC | |
| SBS | Chr3 | 2032542 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g04410 |
| SBS | Chr3 | 29419488 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 29998917 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 13685367 | A-G | HOMO | INTERGENIC | |
| SBS | Chr4 | 22070835 | C-T | HET | INTRON | |
| SBS | Chr4 | 25834734 | C-T | HOMO | UTR_3_PRIME | |
| SBS | Chr5 | 10377689 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 17106249 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g29120 |
| SBS | Chr6 | 24105093 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 21461588 | T-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 23902626 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 27881074 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 27881075 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g46650 |
| SBS | Chr7 | 28002985 | T-C | HET | INTRON | |
| SBS | Chr8 | 17004818 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 2913662 | T-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 18935366 | T-C | HET | INTERGENIC |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 577511 | 577516 | 5 | LOC_Os01g02060 |
| Deletion | Chr3 | 2046957 | 2046960 | 3 | |
| Deletion | Chr1 | 3255489 | 3255499 | 10 | |
| Deletion | Chr5 | 3290122 | 3290149 | 27 | |
| Deletion | Chr2 | 5001895 | 5001896 | 1 | |
| Deletion | Chr9 | 8347164 | 8347173 | 9 | |
| Deletion | Chr8 | 8968715 | 8968716 | 1 | LOC_Os08g14880 |
| Deletion | Chr1 | 9991572 | 9991574 | 2 | LOC_Os01g17360 |
| Deletion | Chr9 | 11031746 | 11034348 | 2602 | LOC_Os09g18020 |
| Deletion | Chr1 | 11216331 | 11216344 | 13 | |
| Deletion | Chr9 | 12640234 | 12640236 | 2 | |
| Deletion | Chr4 | 12977157 | 12977162 | 5 | |
| Deletion | Chr4 | 13562969 | 13562972 | 3 | |
| Deletion | Chr9 | 16995229 | 16995251 | 22 | LOC_Os09g27970 |
| Deletion | Chr5 | 17296001 | 17356000 | 60000 | 11 |
| Deletion | Chr5 | 17427001 | 17520000 | 93000 | 12 |
| Deletion | Chr1 | 17744831 | 17744837 | 6 | |
| Deletion | Chr4 | 18500952 | 18500954 | 2 | |
| Deletion | Chr7 | 18929295 | 18929308 | 13 | LOC_Os07g31850 |
| Deletion | Chr9 | 20112768 | 20112775 | 7 | LOC_Os09g34070 |
| Deletion | Chr6 | 20760651 | 20760658 | 7 | |
| Deletion | Chr7 | 28079548 | 28079563 | 15 | |
| Deletion | Chr11 | 28809496 | 28809497 | 1 | LOC_Os11g47770 |
| Deletion | Chr7 | 28957888 | 28957891 | 3 | LOC_Os07g48420 |
| Deletion | Chr2 | 32222750 | 32222764 | 14 | |
| Deletion | Chr1 | 33168930 | 33168938 | 8 | LOC_Os01g57370 |
| Deletion | Chr1 | 35028266 | 35028273 | 7 | |
| Deletion | Chr1 | 35128175 | 35128182 | 7 | |
| Deletion | Chr1 | 35131919 | 35131922 | 3 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 39558652 | 39558652 | 1 | |
| Insertion | Chr11 | 25763332 | 25763335 | 4 | |
| Insertion | Chr2 | 15646552 | 15646552 | 1 | |
| Insertion | Chr9 | 10403603 | 10403604 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 5668786 | 5767378 | |
| Inversion | Chr9 | 10938087 | 11298647 | LOC_Os09g17880 |
| Inversion | Chr5 | 17295833 | 17425349 | 2 |
| Inversion | Chr5 | 17355916 | 17399474 | LOC_Os05g30070 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 6927431 | Chr7 | 19968670 | LOC_Os11g12410 |
| Translocation | Chr9 | 7646145 | Chr1 | 29297423 | |
| Translocation | Chr9 | 7887765 | Chr4 | 20185757 | |
| Translocation | Chr9 | 7887773 | Chr4 | 20184788 | |
| Translocation | Chr9 | 11320978 | Chr3 | 16003407 | 2 |
| Translocation | Chr9 | 21552131 | Chr2 | 34253001 | LOC_Os09g37310 |
