Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN662-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN662-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 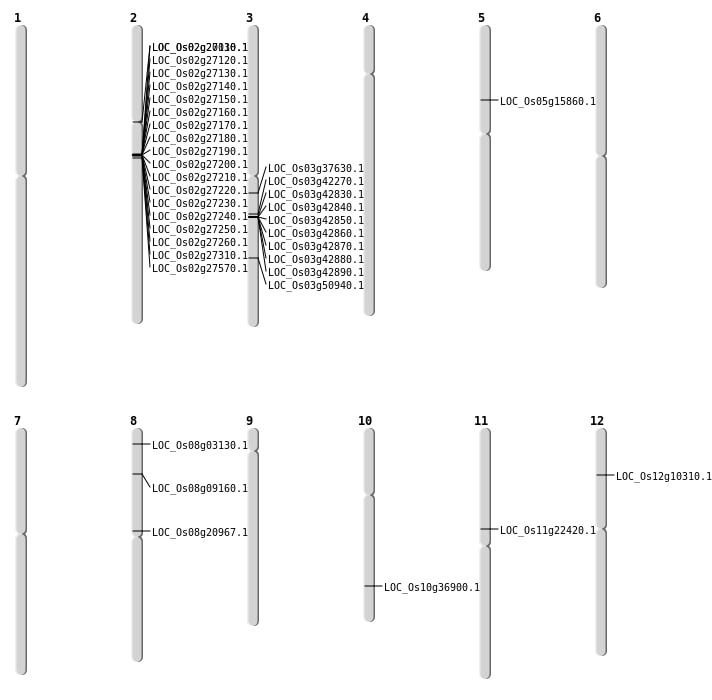
|
| Variant Information |
|---|
Single base substitutions: 19
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18473218 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 4264769 | T-C | HET | INTRON | |
| SBS | Chr11 | 12870979 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g22420 |
| SBS | Chr11 | 13006358 | G-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 10782171 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 11795839 | G-A | HET | STOP_GAINED | LOC_Os02g20030 |
| SBS | Chr3 | 11325068 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 24475507 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 9239304 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 28442261 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 20828193 | A-G | HOMO | INTERGENIC | |
| SBS | Chr5 | 8953739 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g15860 |
| SBS | Chr6 | 25173438 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 6065765 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 12516722 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 20896348 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 9592391 | C-G | HOMO | INTRON | |
| SBS | Chr8 | 1433231 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g03130 |
| SBS | Chr8 | 5311826 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g09160 |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 429498 | 429509 | 11 | |
| Deletion | Chr8 | 1096038 | 1096039 | 1 | |
| Deletion | Chr8 | 2040508 | 2040512 | 4 | |
| Deletion | Chr6 | 2495282 | 2495291 | 9 | |
| Deletion | Chr2 | 4332110 | 4332116 | 6 | |
| Deletion | Chr12 | 5444431 | 5444432 | 1 | LOC_Os12g10310 |
| Deletion | Chr11 | 5860938 | 5860939 | 1 | |
| Deletion | Chr1 | 7665600 | 7665601 | 1 | |
| Deletion | Chr12 | 9229302 | 9229306 | 4 | |
| Deletion | Chr6 | 10879356 | 10879357 | 1 | |
| Deletion | Chr6 | 12539502 | 12539505 | 3 | |
| Deletion | Chr8 | 12558206 | 12558207 | 1 | LOC_Os08g20967 |
| Deletion | Chr1 | 12758428 | 12758434 | 6 | |
| Deletion | Chr9 | 14150143 | 14150144 | 1 | |
| Deletion | Chr2 | 15933001 | 16048000 | 115000 | 16 |
| Deletion | Chr3 | 19185932 | 19185933 | 1 | |
| Deletion | Chr10 | 19776326 | 19777199 | 873 | LOC_Os10g36900 |
| Deletion | Chr8 | 21150875 | 21150878 | 3 | |
| Deletion | Chr2 | 21165831 | 21165842 | 11 | |
| Deletion | Chr3 | 23872001 | 23916000 | 44000 | 7 |
| Deletion | Chr1 | 24828646 | 24828664 | 18 | |
| Deletion | Chr3 | 26210440 | 26210442 | 2 | |
| Deletion | Chr7 | 27801052 | 27801097 | 45 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 8809722 | 8809722 | 1 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 15932996 | 16324519 | 2 |
| Inversion | Chr2 | 16093727 | 16324523 | 2 |
| Inversion | Chr3 | 20867162 | 23517709 | 2 |
| Inversion | Chr3 | 23871807 | 24475557 | LOC_Os03g42830 |
| Inversion | Chr3 | 24475562 | 29136073 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 4726253 | Chr9 | 18723075 | |
| Translocation | Chr12 | 13394797 | Chr3 | 35423623 | |
| Translocation | Chr12 | 21983057 | Chr5 | 12725491 | |
| Translocation | Chr3 | 29085447 | Chr2 | 16047694 | LOC_Os02g27260 |
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr3 | 29101449 | 29101449 | 0 | LOC_Os03g50940 |
