Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN666-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN666-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 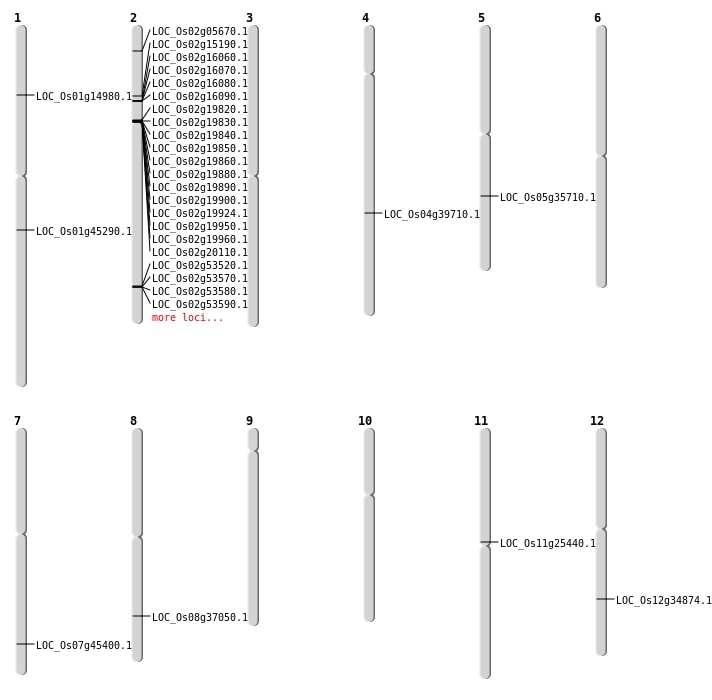
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11759489 | C-G | HET | UTR_3_PRIME | |
| SBS | Chr1 | 19850450 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 2665738 | C-A | HET | INTRON | |
| SBS | Chr1 | 27468427 | C-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 34474204 | T-G | HET | INTRON | |
| SBS | Chr11 | 15180057 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 15427029 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 23420110 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 2452517 | T-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 2452519 | A-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 7665351 | T-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 8362675 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 9751985 | C-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 22811761 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 25522117 | T-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr2 | 475052 | C-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 8470659 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g15190 |
| SBS | Chr2 | 8470660 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 9158504 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g16090 |
| SBS | Chr3 | 13971887 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr4 | 21904148 | A-C | HET | INTRON | |
| SBS | Chr4 | 23665718 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g39710 |
| SBS | Chr5 | 16367145 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 18656884 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 21202454 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g35710 |
| SBS | Chr5 | 27658636 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 14631535 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 15087664 | A-C | HET | INTERGENIC | |
| SBS | Chr6 | 16917794 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 2421030 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr6 | 5533460 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 9081923 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 12482280 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 21865245 | G-C | HOMO | INTRON | |
| SBS | Chr7 | 27080272 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g45400 |
| SBS | Chr7 | 4047211 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 5578623 | A-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 6543809 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 11544627 | A-C | HET | INTERGENIC | |
| SBS | Chr8 | 13480939 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 13480943 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 24789038 | G-A | HOMO | INTERGENIC |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 2505440 | 2505442 | 2 | |
| Deletion | Chr1 | 8397964 | 8397965 | 1 | LOC_Os01g14980 |
| Deletion | Chr2 | 9140001 | 9157000 | 17000 | 4 |
| Deletion | Chr1 | 9982534 | 9982548 | 14 | |
| Deletion | Chr5 | 11348848 | 11348850 | 2 | |
| Deletion | Chr2 | 11584001 | 11747000 | 163000 | 11 |
| Deletion | Chr1 | 12652676 | 12652685 | 9 | |
| Deletion | Chr6 | 14004467 | 14004487 | 20 | |
| Deletion | Chr8 | 20862704 | 20862772 | 68 | |
| Deletion | Chr2 | 21370566 | 21370567 | 1 | |
| Deletion | Chr4 | 21910130 | 21910131 | 1 | |
| Deletion | Chr8 | 22605231 | 22605232 | 1 | |
| Deletion | Chr8 | 23418007 | 23418015 | 8 | LOC_Os08g37050 |
| Deletion | Chr7 | 24144354 | 24144355 | 1 | |
| Deletion | Chr1 | 25711129 | 25711139 | 10 | LOC_Os01g45290 |
| Deletion | Chr11 | 28782639 | 28782641 | 2 | |
| Deletion | Chr2 | 32771280 | 32999437 | 228157 | 33 |
| Deletion | Chr1 | 33239498 | 33239507 | 9 | |
| Deletion | Chr3 | 36197599 | 36197600 | 1 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 12353499 | 12353502 | 4 | |
| Insertion | Chr4 | 9034764 | 9034764 | 1 | |
| Insertion | Chr6 | 23632293 | 23632296 | 4 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 9139951 | 11583748 | LOC_Os02g16060 |
| Inversion | Chr2 | 9156687 | 11850795 | 2 |
| Inversion | Chr2 | 32737958 | 32999434 | LOC_Os02g53520 |
| Inversion | Chr2 | 32737975 | 32771281 | LOC_Os02g53520 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 14501406 | Chr2 | 2772533 | 2 |
| Translocation | Chr12 | 21233040 | Chr1 | 30567628 | LOC_Os12g34874 |
| Translocation | Chr12 | 21236676 | Chr1 | 30567610 |
