Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN671-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN671-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 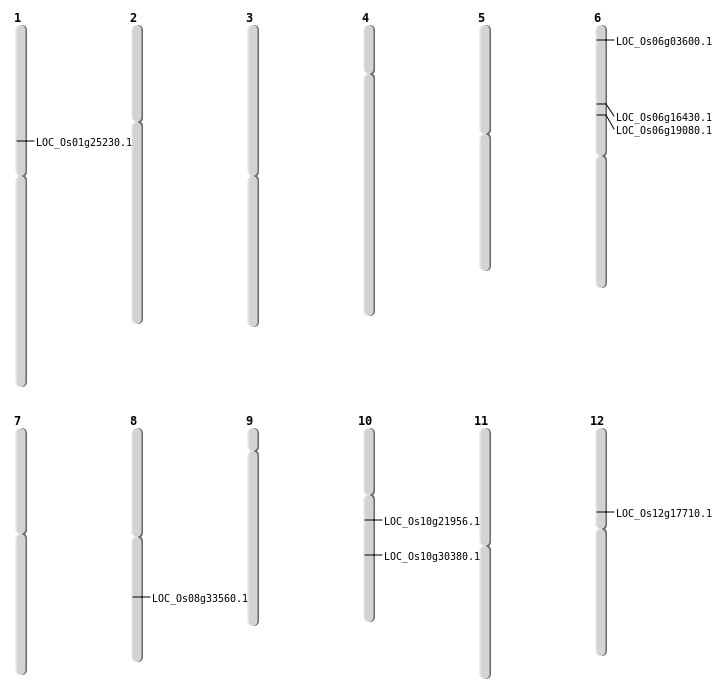
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15821258 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 16829295 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 18221628 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 29997435 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr1 | 8470528 | C-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 15791639 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g30380 |
| SBS | Chr10 | 15791640 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 4061404 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 4061405 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 4061406 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 28924645 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 8042526 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 10147438 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g17710 |
| SBS | Chr12 | 21323821 | T-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr3 | 16447803 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 211046 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 25948172 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 1046544 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 17164097 | T-G | HET | INTERGENIC | |
| SBS | Chr4 | 23606810 | G-C | HOMO | UTR_5_PRIME | |
| SBS | Chr4 | 3657133 | A-G | HET | INTRON | |
| SBS | Chr4 | 9557051 | G-C | HET | INTERGENIC | |
| SBS | Chr5 | 19586440 | T-G | HET | INTERGENIC | |
| SBS | Chr6 | 14667437 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 28516308 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 3097460 | A-T | HET | INTRON | |
| SBS | Chr6 | 9401471 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g16430 |
| SBS | Chr7 | 12911641 | C-G | HOMO | INTRON | |
| SBS | Chr7 | 12911645 | A-G | HOMO | INTRON | |
| SBS | Chr7 | 7596380 | A-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 7596381 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 16609194 | T-G | HET | INTERGENIC | |
| SBS | Chr8 | 3425036 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 21593839 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 4157678 | C-G | HET | INTERGENIC |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 444158 | 444167 | 9 | |
| Deletion | Chr10 | 447028 | 447033 | 5 | |
| Deletion | Chr6 | 1389356 | 1389357 | 1 | LOC_Os06g03600 |
| Deletion | Chr4 | 5902655 | 5902664 | 9 | |
| Deletion | Chr6 | 13123670 | 13123677 | 7 | |
| Deletion | Chr1 | 14251935 | 14251941 | 6 | LOC_Os01g25230 |
| Deletion | Chr6 | 16595491 | 16595494 | 3 | |
| Deletion | Chr12 | 17985318 | 17985322 | 4 | |
| Deletion | Chr11 | 18545136 | 18545149 | 13 | |
| Deletion | Chr7 | 20936657 | 20936662 | 5 | |
| Deletion | Chr8 | 20953367 | 20953386 | 19 | LOC_Os08g33560 |
| Deletion | Chr9 | 20969508 | 20969512 | 4 | |
| Deletion | Chr6 | 21076241 | 21076243 | 2 | |
| Deletion | Chr5 | 22206169 | 22206170 | 1 | |
| Deletion | Chr10 | 22602993 | 22603007 | 14 | |
| Deletion | Chr12 | 24591493 | 24591495 | 2 | |
| Deletion | Chr4 | 26018450 | 26018458 | 8 | |
| Deletion | Chr3 | 28982499 | 28982501 | 2 | |
| Deletion | Chr2 | 29081598 | 29081622 | 24 | |
| Deletion | Chr1 | 37111016 | 37111017 | 1 | |
| Deletion | Chr1 | 37375197 | 37375204 | 7 | |
| Deletion | Chr1 | 42795077 | 42795083 | 6 |
Insertions: 8
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 14409267 | 39977242 | |
| Inversion | Chr1 | 14409283 | 39977009 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 3129575 | Chr6 | 18189917 | |
| Translocation | Chr10 | 11338828 | Chr6 | 10850161 | 2 |
