Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN672-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN672-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 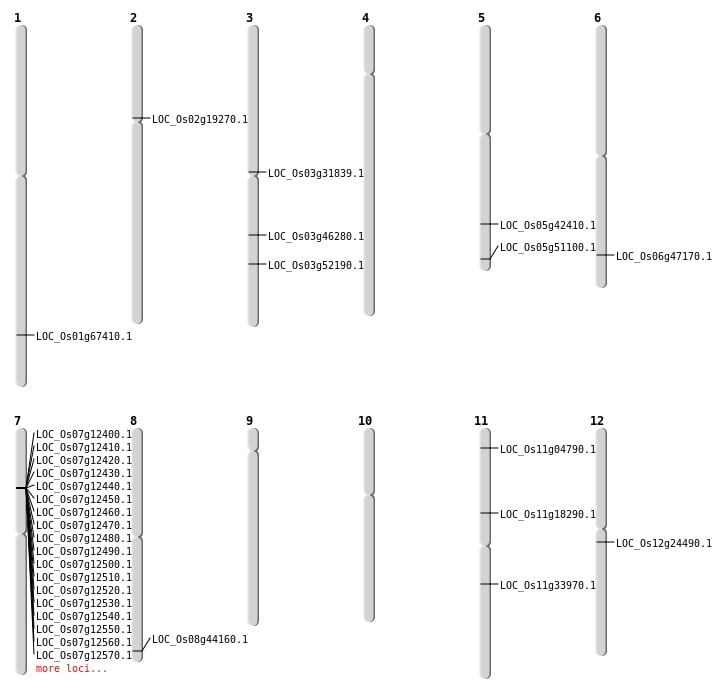
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10321040 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 33758129 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 34864273 | A-G | HOMO | INTRON | |
| SBS | Chr1 | 43085857 | T-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 16607758 | C-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 10306235 | C-T | HET | SPLICE_SITE_ACCEPTOR | LOC_Os11g18290 |
| SBS | Chr11 | 15328066 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 11224151 | C-A | HET | INTRON | |
| SBS | Chr12 | 12948510 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 9918515 | G-T | HET | INTRON | |
| SBS | Chr2 | 10527094 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 11256150 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g19270 |
| SBS | Chr2 | 1142729 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 23618525 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 4973020 | C-T | HET | INTRON | |
| SBS | Chr3 | 19675197 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 26178145 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g46280 |
| SBS | Chr3 | 29963234 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g52190 |
| SBS | Chr3 | 34391226 | C-G | HET | INTERGENIC | |
| SBS | Chr3 | 7753956 | A-T | HET | INTRON | |
| SBS | Chr3 | 9858485 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 7781989 | T-C | HET | INTRON | |
| SBS | Chr5 | 4229558 | C-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 25250331 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 25250332 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 28604747 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os06g47170 |
| SBS | Chr7 | 14335964 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g25100 |
| SBS | Chr7 | 18754503 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 22275493 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 3756915 | A-T | HOMO | INTRON | |
| SBS | Chr7 | 8002786 | T-C | HET | INTRON | |
| SBS | Chr7 | 9413272 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 9413274 | G-C | HET | INTERGENIC | |
| SBS | Chr8 | 24203181 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 18417046 | A-T | HOMO | INTRON | |
| SBS | Chr9 | 7847293 | C-T | HOMO | INTERGENIC |
Deletions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 2044532 | 2044549 | 17 | LOC_Os11g04790 |
| Deletion | Chr7 | 3703481 | 3703483 | 2 | |
| Deletion | Chr5 | 4186327 | 4186334 | 7 | |
| Deletion | Chr7 | 7070001 | 7316000 | 246000 | 39 |
| Deletion | Chr7 | 9431001 | 9602000 | 171000 | 16 |
| Deletion | Chr7 | 9663828 | 9663836 | 8 | LOC_Os07g16490 |
| Deletion | Chr9 | 11598211 | 11598221 | 10 | |
| Deletion | Chr12 | 12400203 | 12400208 | 5 | |
| Deletion | Chr5 | 15693109 | 15693110 | 1 | |
| Deletion | Chr5 | 15706869 | 15706870 | 1 | |
| Deletion | Chr7 | 17196646 | 17196664 | 18 | |
| Deletion | Chr8 | 19354929 | 19354938 | 9 | |
| Deletion | Chr1 | 22215023 | 22215027 | 4 | |
| Deletion | Chr8 | 23926065 | 23926071 | 6 | |
| Deletion | Chr5 | 24838911 | 24839478 | 567 | LOC_Os05g42410 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 39142990 | 39142990 | 1 | LOC_Os01g67410 |
| Insertion | Chr12 | 13982795 | 13982795 | 1 | LOC_Os12g24490 |
| Insertion | Chr2 | 17096141 | 17096143 | 3 | |
| Insertion | Chr4 | 2331262 | 2331263 | 2 | |
| Insertion | Chr5 | 29317059 | 29317060 | 2 | LOC_Os05g51100 |
| Insertion | Chr6 | 7510979 | 7510979 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 7070115 | 9430936 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 16323335 | Chr2 | 8190350 | |
| Translocation | Chr3 | 18216592 | Chr1 | 9105546 | LOC_Os03g31839 |
| Translocation | Chr10 | 19292221 | Chr3 | 31080052 | |
| Translocation | Chr11 | 19861965 | Chr7 | 7093264 | 2 |
| Translocation | Chr8 | 27802443 | Chr5 | 24839472 | 2 |
| Translocation | Chr8 | 27802448 | Chr5 | 24838923 | LOC_Os08g44160 |
