Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN673-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN673-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 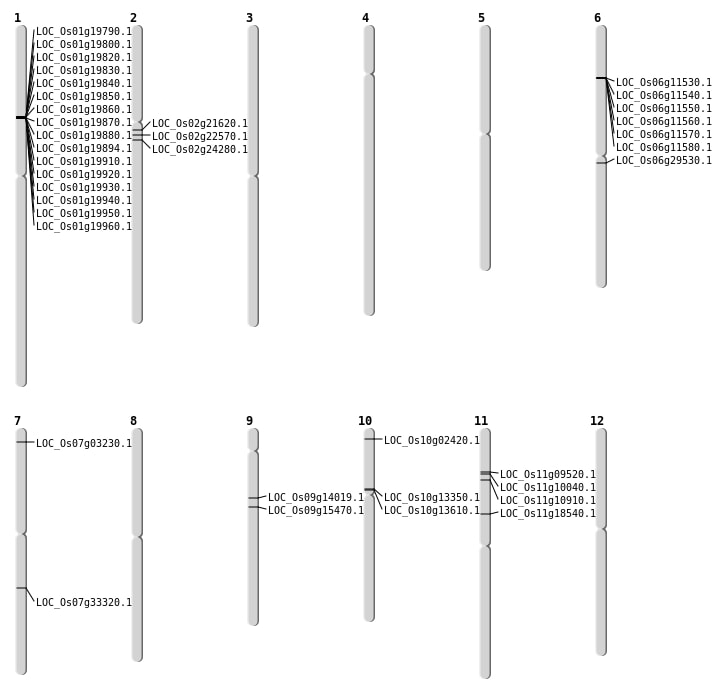
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21079068 | C-A | HOMO | INTRON | |
| SBS | Chr10 | 10695854 | T-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 7209217 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os10g13350 |
| SBS | Chr10 | 7380444 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g13610 |
| SBS | Chr10 | 885369 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g02420 |
| SBS | Chr11 | 11183588 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 20300916 | A-T | HOMO | INTRON | |
| SBS | Chr11 | 7391556 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 10844427 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 19447421 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 8199901 | A-T | HET | INTRON | |
| SBS | Chr2 | 14082178 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g24280 |
| SBS | Chr2 | 23962042 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 29972400 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 348482 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 348483 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 6376474 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 33122697 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 9704988 | G-T | HOMO | INTRON | |
| SBS | Chr4 | 26826213 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 11946045 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 14557293 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 14557294 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 20387783 | T-C | HOMO | INTRON | |
| SBS | Chr5 | 20387786 | G-T | HOMO | INTRON | |
| SBS | Chr5 | 20387787 | T-A | HOMO | INTRON | |
| SBS | Chr6 | 10588247 | G-A | HOMO | UTR_3_PRIME | |
| SBS | Chr6 | 12981940 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 21420894 | A-T | HOMO | INTRON | |
| SBS | Chr7 | 26752557 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 8475091 | A-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 24530469 | A-C | HET | INTERGENIC | |
| SBS | Chr8 | 28287040 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 3804896 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 22541764 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 2569825 | G-A | HET | INTRON | |
| SBS | Chr9 | 5679972 | C-T | HET | INTERGENIC |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 18738 | 18739 | 1 | |
| Deletion | Chr9 | 1307250 | 1307253 | 3 | |
| Deletion | Chr5 | 1509716 | 1509720 | 4 | |
| Deletion | Chr9 | 4672890 | 4672906 | 16 | |
| Deletion | Chr6 | 6112796 | 6141802 | 29006 | 6 |
| Deletion | Chr2 | 7008748 | 7008756 | 8 | |
| Deletion | Chr7 | 7408300 | 7408303 | 3 | |
| Deletion | Chr9 | 8284888 | 8284895 | 7 | LOC_Os09g14019 |
| Deletion | Chr7 | 9193636 | 9193648 | 12 | |
| Deletion | Chr9 | 9468170 | 9468171 | 1 | LOC_Os09g15470 |
| Deletion | Chr1 | 11225001 | 11325000 | 100000 | 15 |
| Deletion | Chr1 | 13932640 | 13932648 | 8 | |
| Deletion | Chr2 | 14060964 | 14060965 | 1 | |
| Deletion | Chr1 | 14108517 | 14108542 | 25 | |
| Deletion | Chr5 | 14563335 | 14563348 | 13 | |
| Deletion | Chr11 | 15169118 | 15169809 | 691 | |
| Deletion | Chr5 | 15309014 | 15309015 | 1 | |
| Deletion | Chr7 | 19026030 | 19026032 | 2 | |
| Deletion | Chr7 | 19917077 | 19917090 | 13 | LOC_Os07g33320 |
| Deletion | Chr10 | 21565482 | 21565484 | 2 | |
| Deletion | Chr10 | 23024022 | 23024025 | 3 | |
| Deletion | Chr8 | 25123448 | 25123456 | 8 | |
| Deletion | Chr4 | 26640872 | 26640874 | 2 | |
| Deletion | Chr1 | 29039332 | 29039336 | 4 | |
| Deletion | Chr4 | 29109866 | 29109874 | 8 |
Insertions: 11
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 23798371 | 23798372 | 2 | |
| Insertion | Chr1 | 40468997 | 40468999 | 3 | |
| Insertion | Chr11 | 22634284 | 22634285 | 2 | |
| Insertion | Chr11 | 5372649 | 5372656 | 8 | LOC_Os11g10040 |
| Insertion | Chr2 | 17969513 | 17969514 | 2 | |
| Insertion | Chr3 | 25745703 | 25745705 | 3 | |
| Insertion | Chr7 | 1274621 | 1274644 | 24 | LOC_Os07g03230 |
| Insertion | Chr7 | 1455532 | 1455543 | 12 | |
| Insertion | Chr7 | 24313236 | 24313237 | 2 | |
| Insertion | Chr8 | 20289288 | 20289290 | 3 | |
| Insertion | Chr9 | 2166626 | 2166657 | 32 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 6023183 | 6188754 | LOC_Os11g10910 |
| Inversion | Chr1 | 9383903 | 13918587 | |
| Inversion | Chr1 | 11190282 | 13918447 | |
| Inversion | Chr2 | 12153563 | 12844308 | LOC_Os02g21620 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 4352543 | Chr1 | 11324341 | |
| Translocation | Chr11 | 5104116 | Chr6 | 16925173 | 2 |
| Translocation | Chr11 | 6023184 | Chr1 | 11333891 | 2 |
| Translocation | Chr11 | 6188756 | Chr1 | 11324809 | |
| Translocation | Chr11 | 10449195 | Chr6 | 17476343 | LOC_Os11g18540 |
| Translocation | Chr10 | 13748729 | Chr2 | 13460116 | LOC_Os02g22570 |
| Translocation | Chr6 | 18200621 | Chr2 | 8950278 |
