Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN692-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN692-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 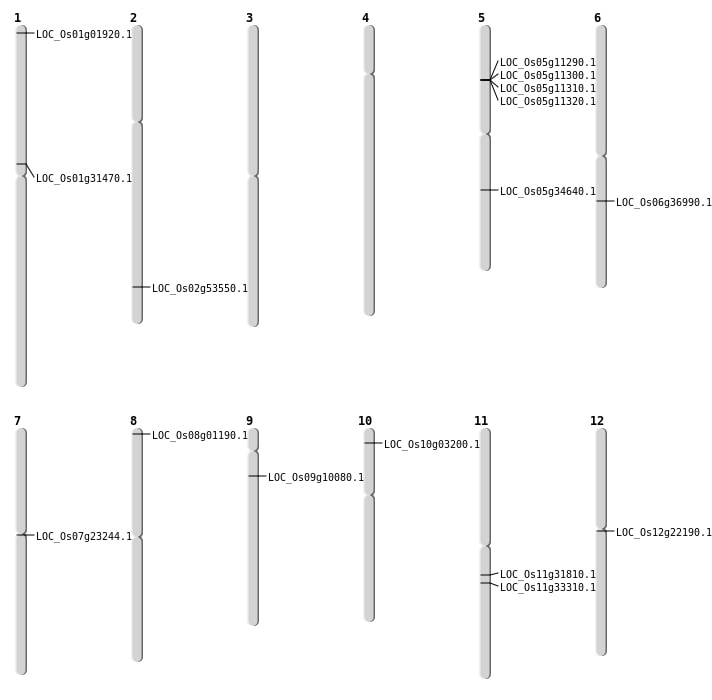
|
| Variant Information |
|---|
Single base substitutions: 42
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14928894 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 17833819 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 20335370 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 23771899 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 24985921 | C-G | HET | INTERGENIC | |
| SBS | Chr1 | 36517071 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 1353092 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g03200 |
| SBS | Chr10 | 18618366 | C-T | HOMO | INTRON | |
| SBS | Chr10 | 18618367 | A-T | HOMO | INTRON | |
| SBS | Chr11 | 11477660 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 133525 | T-C | HOMO | INTRON | |
| SBS | Chr11 | 22879613 | G-A | HET | INTRON | |
| SBS | Chr11 | 4784101 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 10271759 | G-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 12500361 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 12500362 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g22190 |
| SBS | Chr12 | 12922767 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 25900663 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 25926034 | C-T | HOMO | INTRON | |
| SBS | Chr2 | 2791140 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 32764606 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g53550 |
| SBS | Chr2 | 3598467 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 17003714 | T-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 17643327 | G-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 2796346 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 3088662 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 10955733 | A-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 10856288 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 13998357 | G-A | HET | INTRON | |
| SBS | Chr5 | 17553893 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 19884892 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 20543683 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g34640 |
| SBS | Chr6 | 16196322 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 13005329 | T-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr7 | 3623912 | C-A | HOMO | INTRON | |
| SBS | Chr8 | 11649009 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 17695018 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 19355242 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 8934050 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 8934051 | T-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 3618784 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 5495450 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g10080 |
Deletions: 18
Insertions: 5
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 1919998 | 19701657 | LOC_Os11g33310 |
| Inversion | Chr1 | 17214942 | 17242957 | LOC_Os01g31470 |
| Inversion | Chr1 | 17215524 | 17242960 | LOC_Os01g31470 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 138559 | Chr1 | 17215506 | 2 |
| Translocation | Chr8 | 138570 | Chr1 | 502159 | 2 |
| Translocation | Chr7 | 226868 | Chr2 | 21806144 | |
| Translocation | Chr7 | 13101031 | Chr6 | 21809536 | 2 |
| Translocation | Chr11 | 18672151 | Chr7 | 4798108 | LOC_Os11g31810 |
| Translocation | Chr10 | 19398843 | Chr8 | 19810524 | |
| Translocation | Chr5 | 22882401 | Chr1 | 545019 | |
| Translocation | Chr5 | 22882414 | Chr1 | 544784 |
