Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN695-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN695-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 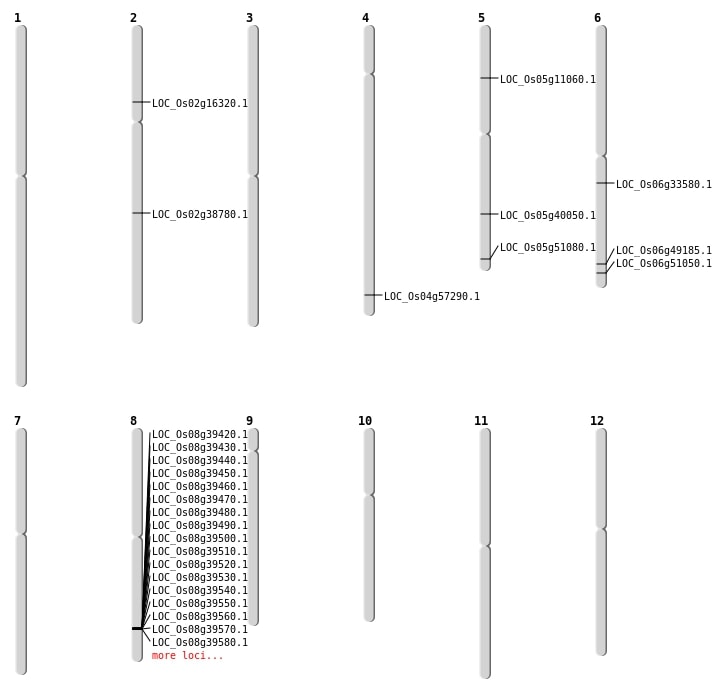
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11186571 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 28805962 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 3986488 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 6955497 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 5277175 | C-A | HOMO | INTRON | |
| SBS | Chr12 | 10525984 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 10525985 | T-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 27445063 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 10238283 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 21094027 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 9280587 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g16320 |
| SBS | Chr3 | 20054016 | G-A | HOMO | INTRON | |
| SBS | Chr3 | 21553602 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 14289248 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 23526877 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g40050 |
| SBS | Chr5 | 24399807 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 29312305 | G-A | HET | STOP_GAINED | LOC_Os05g51080 |
| SBS | Chr5 | 6193515 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g11060 |
| SBS | Chr6 | 10794688 | C-T | HET | INTRON | |
| SBS | Chr6 | 22427494 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 23163144 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 29803213 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g49185 |
| SBS | Chr6 | 30887872 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g51050 |
| SBS | Chr7 | 1445595 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 14811812 | A-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 12181242 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 24257743 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 24257745 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 27459081 | C-T | HET | INTRON | |
| SBS | Chr8 | 6885407 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 7524565 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 7524566 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 9581802 | A-T | HET | INTERGENIC |
Deletions: 7
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 9534458 | 9534464 | 6 | |
| Deletion | Chr12 | 11745074 | 11745075 | 1 | |
| Deletion | Chr7 | 14811813 | 14811814 | 1 | |
| Deletion | Chr6 | 19555015 | 19555022 | 7 | LOC_Os06g33580 |
| Deletion | Chr8 | 24928001 | 25118000 | 190000 | 27 |
| Deletion | Chr3 | 29097388 | 29097394 | 6 | |
| Deletion | Chr4 | 34111435 | 34111442 | 7 | LOC_Os04g57290 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 26163275 | 26163277 | 3 | |
| Insertion | Chr9 | 11342421 | 11342422 | 2 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 14550621 | 14709990 | |
| Inversion | Chr1 | 14709997 | 14954737 | |
| Inversion | Chr1 | 16912069 | 16949155 | |
| Inversion | Chr2 | 23441741 | 23963874 | LOC_Os02g38780 |
| Inversion | Chr12 | 24982339 | 25044363 |
No Translocation
