Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN701-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN701-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 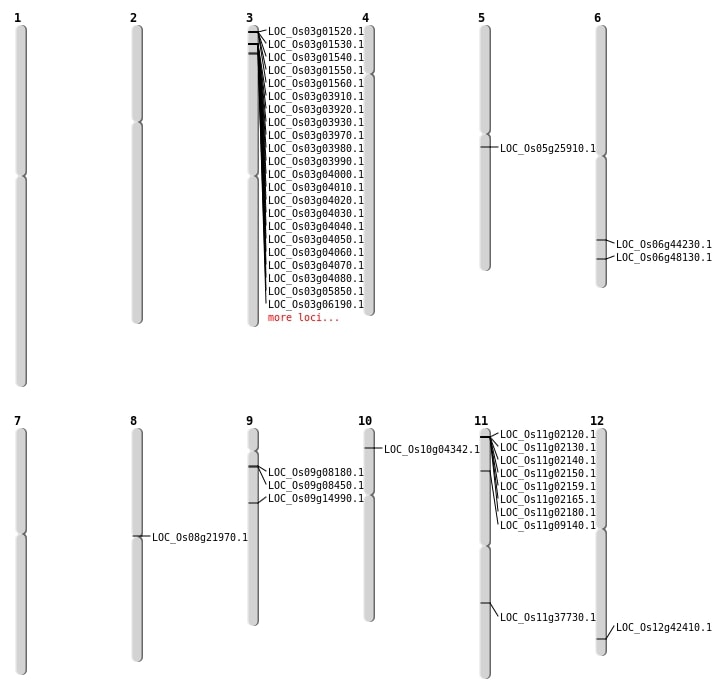
|
| Variant Information |
|---|
Single base substitutions: 40
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10730299 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 40672743 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 6850716 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 15725851 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 20993200 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 22309303 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g37730 |
| SBS | Chr11 | 28626838 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 580294 | A-G | HOMO | INTRON | |
| SBS | Chr12 | 16052271 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr12 | 26910928 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 7397489 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 12750398 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 15182097 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 16294550 | A-C | HET | INTRON | |
| SBS | Chr2 | 26368314 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 31949815 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 3726160 | C-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 1020052 | G-C | HET | UTR_3_PRIME | |
| SBS | Chr3 | 17944360 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 2939611 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g05850 |
| SBS | Chr3 | 32858907 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 34231871 | T-C | HOMO | INTRON | |
| SBS | Chr3 | 5271831 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g10334 |
| SBS | Chr4 | 15930417 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 7721802 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 10110812 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 12227012 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 15069844 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g25910 |
| SBS | Chr5 | 24586870 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 26682959 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g44230 |
| SBS | Chr8 | 10407652 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 13168110 | T-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g21970 |
| SBS | Chr8 | 19170691 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 26503517 | T-A | HET | INTRON | |
| SBS | Chr8 | 4081625 | G-T | HOMO | INTRON | |
| SBS | Chr9 | 4236149 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g08180 |
| SBS | Chr9 | 4236150 | A-T | HOMO | STOP_GAINED | LOC_Os09g08180 |
| SBS | Chr9 | 4236152 | A-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g08180 |
| SBS | Chr9 | 8996989 | T-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr9 | 8996990 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g14990 |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 103893 | 103904 | 11 | |
| Deletion | Chr9 | 130262 | 130289 | 27 | |
| Deletion | Chr3 | 327226 | 356502 | 29276 | 5 |
| Deletion | Chr11 | 579565 | 607034 | 27469 | 7 |
| Deletion | Chr3 | 1781001 | 1871000 | 90000 | 16 |
| Deletion | Chr5 | 2164752 | 2164753 | 1 | |
| Deletion | Chr8 | 2465191 | 2465197 | 6 | |
| Deletion | Chr3 | 3109020 | 3109033 | 13 | LOC_Os03g06190 |
| Deletion | Chr8 | 3194768 | 3194770 | 2 | |
| Deletion | Chr10 | 3629691 | 3629959 | 268 | |
| Deletion | Chr9 | 4408736 | 4408746 | 10 | LOC_Os09g08450 |
| Deletion | Chr2 | 5722189 | 5722191 | 2 | |
| Deletion | Chr4 | 8341138 | 8341139 | 1 | |
| Deletion | Chr7 | 11700349 | 11700353 | 4 | |
| Deletion | Chr7 | 11758300 | 11758303 | 3 | |
| Deletion | Chr5 | 16351030 | 16351031 | 1 | |
| Deletion | Chr8 | 17862076 | 17862082 | 6 | |
| Deletion | Chr7 | 22644319 | 22644324 | 5 | |
| Deletion | Chr9 | 22769660 | 22769661 | 1 | |
| Deletion | Chr6 | 24618315 | 24618319 | 4 | |
| Deletion | Chr8 | 24622550 | 24622551 | 1 | |
| Deletion | Chr11 | 25679441 | 25679449 | 8 | |
| Deletion | Chr12 | 26359369 | 26359381 | 12 | LOC_Os12g42410 |
| Deletion | Chr7 | 26609590 | 26609595 | 5 | |
| Deletion | Chr8 | 27371640 | 27371649 | 9 | |
| Deletion | Chr6 | 29120764 | 29120815 | 51 | LOC_Os06g48130 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 4866717 | 4866718 | 2 | LOC_Os11g09140 |
| Insertion | Chr2 | 18025170 | 18025171 | 2 | |
| Insertion | Chr3 | 16124928 | 16124928 | 1 | |
| Insertion | Chr4 | 34768945 | 34768945 | 1 |
No Inversion
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 2044495 | Chr4 | 12757392 | LOC_Os10g04342 |
| Translocation | Chr11 | 3435043 | Chr10 | 466399 | |
| Translocation | Chr11 | 22938433 | Chr6 | 1671809 |
