Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN704-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN704-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Phenotype Changes (Hover to Zoom-In) |  |
| Mapping (Hover to Zoom-In) | 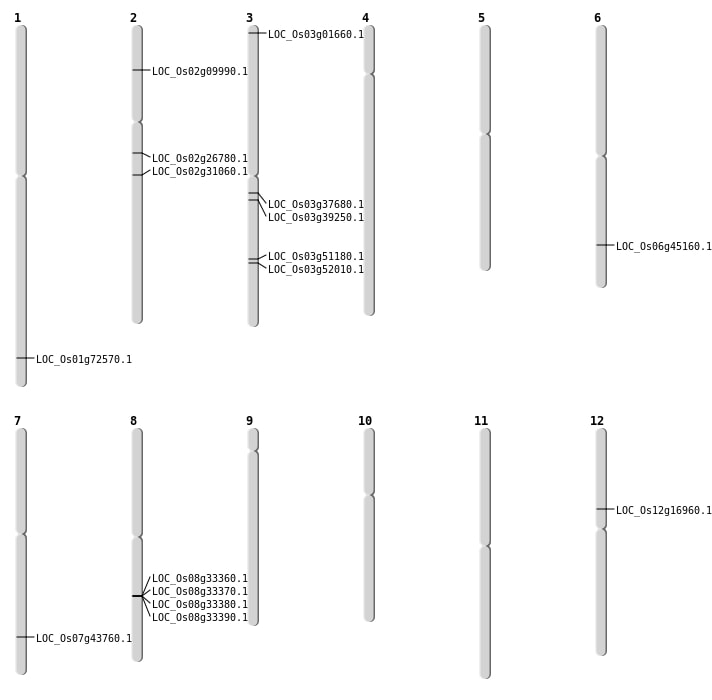
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 43024685 | A-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 6998763 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 982596 | A-G | HOMO | INTERGENIC | |
| SBS | Chr12 | 20308857 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 7766142 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 8985179 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 9715528 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g16960 |
| SBS | Chr2 | 10102959 | C-T | HOMO | INTRON | |
| SBS | Chr2 | 3698170 | A-C | HET | INTRON | |
| SBS | Chr2 | 3971653 | G-T | HET | UTR_5_PRIME | |
| SBS | Chr2 | 4090175 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 6359217 | G-C | HOMO | INTERGENIC | |
| SBS | Chr3 | 13262125 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 19992793 | A-C | HET | INTERGENIC | |
| SBS | Chr3 | 20791399 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 29285306 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g51180 |
| SBS | Chr3 | 29845295 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g52010 |
| SBS | Chr3 | 407405 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g01660 |
| SBS | Chr3 | 4467224 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 24127913 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 28462886 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 477554 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 4879746 | C-T | HOMO | INTRON | |
| SBS | Chr5 | 767276 | A-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 1757929 | A-T | HOMO | INTRON | |
| SBS | Chr6 | 30996592 | A-G | HOMO | INTERGENIC | |
| SBS | Chr6 | 6451998 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 26185455 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g43760 |
| SBS | Chr7 | 9981805 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 27404700 | C-T | HOMO | INTRON | |
| SBS | Chr8 | 9071121 | G-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 19954683 | T-C | HET | INTERGENIC |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 226905 | 226907 | 2 | |
| Deletion | Chr5 | 766225 | 766229 | 4 | |
| Deletion | Chr7 | 1250582 | 1250587 | 5 | |
| Deletion | Chr10 | 2277007 | 2277072 | 65 | |
| Deletion | Chr2 | 5194495 | 5194496 | 1 | LOC_Os02g09990 |
| Deletion | Chr3 | 9267422 | 9267435 | 13 | |
| Deletion | Chr8 | 12784640 | 12784641 | 1 | |
| Deletion | Chr10 | 13106376 | 13106380 | 4 | |
| Deletion | Chr7 | 14304461 | 14304475 | 14 | |
| Deletion | Chr10 | 20269765 | 20269831 | 66 | |
| Deletion | Chr8 | 20810001 | 20827000 | 17000 | 4 |
| Deletion | Chr6 | 20907260 | 20907261 | 1 | |
| Deletion | Chr3 | 21810630 | 21810639 | 9 | LOC_Os03g39250 |
| Deletion | Chr9 | 21916144 | 21916149 | 5 | |
| Deletion | Chr2 | 23636259 | 23636270 | 11 | |
| Deletion | Chr6 | 25903367 | 25903368 | 1 | |
| Deletion | Chr6 | 27307127 | 27307128 | 1 | LOC_Os06g45160 |
| Deletion | Chr11 | 27677819 | 27677824 | 5 | |
| Deletion | Chr2 | 29308827 | 29308830 | 3 | |
| Deletion | Chr3 | 33624258 | 33624275 | 17 | |
| Deletion | Chr1 | 42095294 | 42095296 | 2 | LOC_Os01g72570 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 42774203 | 42774204 | 2 | |
| Insertion | Chr2 | 18556721 | 18556726 | 6 | LOC_Os02g31060 |
| Insertion | Chr3 | 22561110 | 22561111 | 2 | |
| Insertion | Chr5 | 2339784 | 2339784 | 1 | |
| Insertion | Chr9 | 14806864 | 14806865 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 350482 | 763834 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 6589435 | Chr3 | 20898018 | LOC_Os03g37680 |
| Translocation | Chr9 | 6590096 | Chr3 | 20898017 | LOC_Os03g37680 |
| Translocation | Chr9 | 6590107 | Chr5 | 763853 | |
| Translocation | Chr9 | 6590841 | Chr5 | 350483 | |
| Translocation | Chr8 | 11601792 | Chr4 | 61845 | |
| Translocation | Chr2 | 15723259 | Chr1 | 9625506 | LOC_Os02g26780 |
| Translocation | Chr11 | 22565555 | Chr9 | 8109240 |
