Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN711-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN711-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 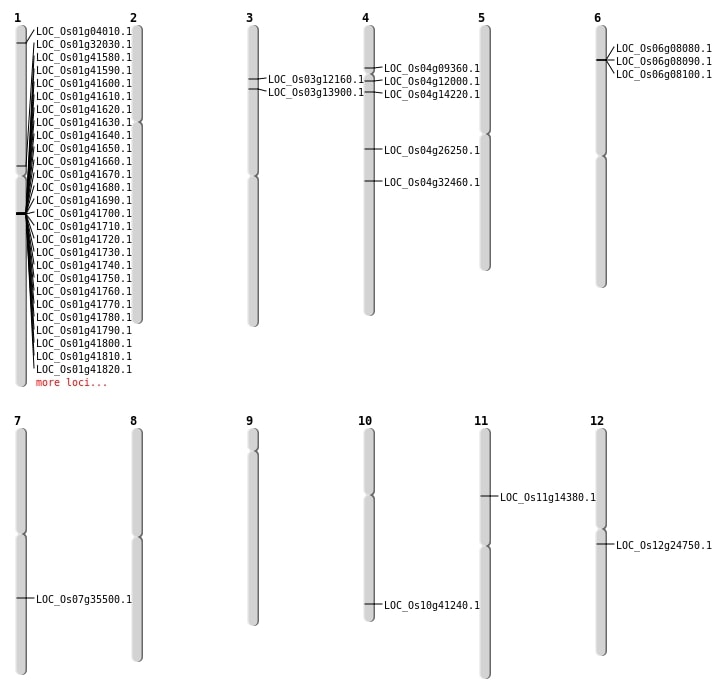
|
| Variant Information |
|---|
Single base substitutions: 30
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 25247238 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 26328581 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 3393632 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 34990975 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 41206915 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g71220 |
| SBS | Chr10 | 17345168 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 17301569 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 19247396 | C-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 14200120 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g24750 |
| SBS | Chr12 | 20875092 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 6306913 | G-A | HET | INTRON | |
| SBS | Chr2 | 8987635 | A-T | HET | UTR_5_PRIME | |
| SBS | Chr3 | 11912972 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 6383840 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g12160 |
| SBS | Chr3 | 7538188 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g13900 |
| SBS | Chr4 | 10275190 | T-A | HET | INTRON | |
| SBS | Chr4 | 15915452 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 23215951 | C-T | HET | INTRON | |
| SBS | Chr4 | 4983606 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g09360 |
| SBS | Chr5 | 11749525 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 14911797 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 16934256 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 29891655 | A-T | HET | INTRON | |
| SBS | Chr7 | 20783800 | G-A | HOMO | INTRON | |
| SBS | Chr8 | 10583423 | C-G | HET | INTERGENIC | |
| SBS | Chr8 | 20692824 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 20692825 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 14690705 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 5496117 | A-C | HET | INTERGENIC | |
| SBS | Chr9 | 981317 | C-T | HET | INTERGENIC |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 1483849 | 1483854 | 5 | |
| Deletion | Chr1 | 1739214 | 1739215 | 1 | LOC_Os01g04010 |
| Deletion | Chr4 | 2273232 | 2273237 | 5 | |
| Deletion | Chr7 | 3143254 | 3143267 | 13 | |
| Deletion | Chr6 | 3921001 | 3933000 | 12000 | 3 |
| Deletion | Chr2 | 6299542 | 6299543 | 1 | |
| Deletion | Chr11 | 8073240 | 8073252 | 12 | LOC_Os11g14380 |
| Deletion | Chr4 | 9401837 | 9401838 | 1 | |
| Deletion | Chr6 | 10784913 | 10784914 | 1 | |
| Deletion | Chr4 | 12723899 | 12723908 | 9 | |
| Deletion | Chr11 | 12924779 | 12924782 | 3 | |
| Deletion | Chr5 | 13287153 | 13287156 | 3 | |
| Deletion | Chr4 | 15311624 | 15311633 | 9 | LOC_Os04g26250 |
| Deletion | Chr2 | 18990679 | 18990692 | 13 | |
| Deletion | Chr10 | 18993078 | 18993081 | 3 | |
| Deletion | Chr4 | 19457354 | 19457359 | 5 | LOC_Os04g32460 |
| Deletion | Chr2 | 20262115 | 20262117 | 2 | |
| Deletion | Chr10 | 22162172 | 22162178 | 6 | LOC_Os10g41240 |
| Deletion | Chr1 | 23533533 | 23989928 | 456395 | 68 |
| Deletion | Chr5 | 26131319 | 26131344 | 25 | |
| Deletion | Chr3 | 32778005 | 32778007 | 2 | |
| Deletion | Chr1 | 33208924 | 33208925 | 1 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr5 | 6179376 | 6179377 | 2 | |
| Insertion | Chr6 | 12066029 | 12066029 | 1 | |
| Insertion | Chr6 | 27435499 | 27435499 | 1 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 6585867 | 6588953 | LOC_Os04g12000 |
| Inversion | Chr4 | 6588976 | 7980584 | 2 |
| Inversion | Chr8 | 12181573 | 13781862 | |
| Inversion | Chr8 | 12181589 | 13781870 | |
| Inversion | Chr1 | 17167806 | 17535659 | LOC_Os01g32030 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 5024481 | Chr3 | 31077495 | |
| Translocation | Chr7 | 21235194 | Chr4 | 6585797 | LOC_Os07g35500 |
