Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN712-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN712-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 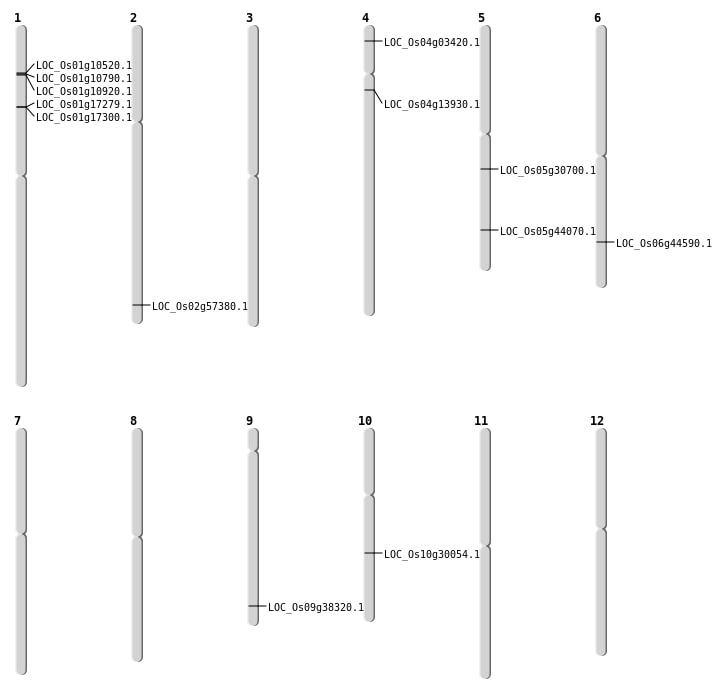
|
| Variant Information |
|---|
Single base substitutions: 29
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 7473342 | T-C | HET | INTERGENIC | |
| SBS | Chr10 | 1742161 | G-A | HET | INTRON | |
| SBS | Chr10 | 253726 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 3437519 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 3303753 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 10491810 | A-G | HET | INTRON | |
| SBS | Chr12 | 693130 | C-T | HOMO | INTRON | |
| SBS | Chr2 | 11323901 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 14870314 | A-T | HOMO | SPLICE_SITE_REGION | |
| SBS | Chr2 | 23559698 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 35160086 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g57380 |
| SBS | Chr2 | 35297033 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 35331126 | T-C | HET | INTRON | |
| SBS | Chr3 | 32071746 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 3841589 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 1468057 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g03420 |
| SBS | Chr4 | 17524063 | C-T | HET | INTRON | |
| SBS | Chr4 | 30613890 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 7781595 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g13930 |
| SBS | Chr5 | 18025599 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 25625627 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g44070 |
| SBS | Chr6 | 14654524 | C-T | HOMO | INTRON | |
| SBS | Chr7 | 15151600 | G-A | HET | INTRON | |
| SBS | Chr7 | 26496780 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 20725407 | G-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 28036660 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 28344444 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 5947716 | T-C | HET | UTR_5_PRIME | |
| SBS | Chr9 | 2745806 | A-T | HET | INTERGENIC |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 2411145 | 2411146 | 1 | |
| Deletion | Chr9 | 4738257 | 4738271 | 14 | |
| Deletion | Chr6 | 5333293 | 5333297 | 4 | |
| Deletion | Chr2 | 5539085 | 5539098 | 13 | |
| Deletion | Chr5 | 6159240 | 6159243 | 3 | |
| Deletion | Chr7 | 6218090 | 6218093 | 3 | |
| Deletion | Chr11 | 6642110 | 6642125 | 15 | |
| Deletion | Chr1 | 9943001 | 9956000 | 13000 | 2 |
| Deletion | Chr6 | 10191381 | 10191382 | 1 | |
| Deletion | Chr5 | 14213648 | 14213650 | 2 | |
| Deletion | Chr4 | 14746035 | 14746073 | 38 | |
| Deletion | Chr1 | 15460691 | 15460694 | 3 | |
| Deletion | Chr4 | 15529287 | 15529298 | 11 | |
| Deletion | Chr10 | 15601765 | 15601767 | 2 | LOC_Os10g30054 |
| Deletion | Chr5 | 16410486 | 16410501 | 15 | |
| Deletion | Chr11 | 17770003 | 17770018 | 15 | |
| Deletion | Chr5 | 17780367 | 17780371 | 4 | LOC_Os05g30700 |
| Deletion | Chr5 | 17852239 | 17852259 | 20 | |
| Deletion | Chr1 | 21349814 | 21349819 | 5 | |
| Deletion | Chr8 | 21559220 | 21559221 | 1 | |
| Deletion | Chr9 | 22042663 | 22042685 | 22 | LOC_Os09g38320 |
| Deletion | Chr2 | 23923687 | 23923695 | 8 | |
| Deletion | Chr6 | 26925753 | 26925773 | 20 | LOC_Os06g44590 |
| Deletion | Chr6 | 29195208 | 29195214 | 6 | |
| Deletion | Chr1 | 30705742 | 30705744 | 2 |
Insertions: 5
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 5583112 | 5763039 | 2 |
| Inversion | Chr1 | 5763043 | 5826530 | 2 |
| Inversion | Chr2 | 33652756 | 35680031 | |
| Inversion | Chr2 | 33652816 | 35680032 |
No Translocation
