Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN713-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN713-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 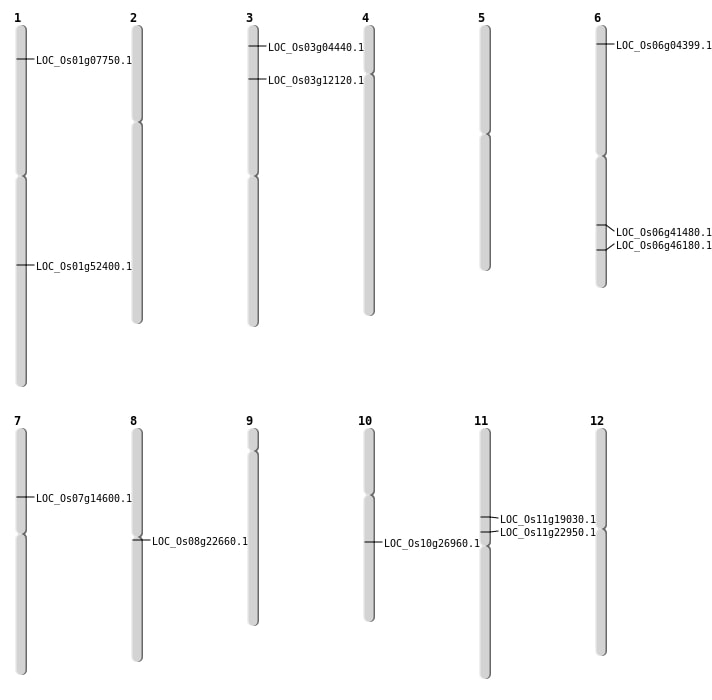
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16156299 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 28575831 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr1 | 29824040 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 30711493 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 40538347 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 8604426 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 16464903 | C-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 3378812 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 13194112 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g22950 |
| SBS | Chr11 | 20922939 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 8430313 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 27487980 | C-T | HET | INTRON | |
| SBS | Chr2 | 12881435 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 30118310 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 30118311 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 30385093 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 17243360 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 2061986 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g04440 |
| SBS | Chr3 | 28634153 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 2893819 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 29988826 | G-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 2004108 | C-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 24180913 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 28275783 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 5841851 | T-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 8869297 | T-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 26338811 | A-G | HET | INTRON | |
| SBS | Chr7 | 8326870 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os07g14600 |
| SBS | Chr8 | 13633940 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g22660 |
| SBS | Chr8 | 14522525 | C-T | HOMO | INTRON | |
| SBS | Chr8 | 16595349 | C-T | HOMO | INTRON | |
| SBS | Chr8 | 3030420 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 5975918 | T-C | HOMO | INTERGENIC | |
| SBS | Chr9 | 10164374 | T-G | HET | INTERGENIC | |
| SBS | Chr9 | 11750154 | T-C | HET | INTERGENIC |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 219574 | 219577 | 3 | |
| Deletion | Chr1 | 1103135 | 1103139 | 4 | |
| Deletion | Chr8 | 2106827 | 2106829 | 2 | |
| Deletion | Chr1 | 3721275 | 3721304 | 29 | LOC_Os01g07750 |
| Deletion | Chr3 | 6363054 | 6363059 | 5 | LOC_Os03g12120 |
| Deletion | Chr11 | 10852930 | 10852931 | 1 | LOC_Os11g19030 |
| Deletion | Chr10 | 11141367 | 11141374 | 7 | |
| Deletion | Chr11 | 13972953 | 13972973 | 20 | |
| Deletion | Chr10 | 14197741 | 14197751 | 10 | LOC_Os10g26960 |
| Deletion | Chr5 | 16384435 | 16384452 | 17 | |
| Deletion | Chr1 | 18222604 | 18222610 | 6 | |
| Deletion | Chr8 | 19176710 | 19176713 | 3 | |
| Deletion | Chr12 | 21609945 | 21609964 | 19 | |
| Deletion | Chr1 | 21657724 | 21657746 | 22 | |
| Deletion | Chr5 | 21817496 | 21817503 | 7 | |
| Deletion | Chr10 | 23087741 | 23087742 | 1 | |
| Deletion | Chr6 | 24868173 | 24868174 | 1 | LOC_Os06g41480 |
| Deletion | Chr6 | 27055631 | 27055632 | 1 | |
| Deletion | Chr1 | 27251249 | 27251296 | 47 | |
| Deletion | Chr6 | 27791332 | 27791432 | 100 | |
| Deletion | Chr6 | 27990540 | 27990550 | 10 | LOC_Os06g46180 |
| Deletion | Chr1 | 30106375 | 30106376 | 1 | LOC_Os01g52400 |
| Deletion | Chr2 | 32223953 | 32223954 | 1 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 4878367 | 4878368 | 2 | |
| Insertion | Chr4 | 20701596 | 20701597 | 2 | |
| Insertion | Chr7 | 11316860 | 11316866 | 7 | |
| Insertion | Chr7 | 1525156 | 1525156 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 36312464 | 36603065 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 9639766 | Chr6 | 1887417 | LOC_Os06g04399 |
| Translocation | Chr11 | 17698153 | Chr5 | 22429614 | |
| Translocation | Chr5 | 17992514 | Chr1 | 36603071 | |
| Translocation | Chr5 | 18007202 | Chr1 | 36312470 |
