Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN714-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN714-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 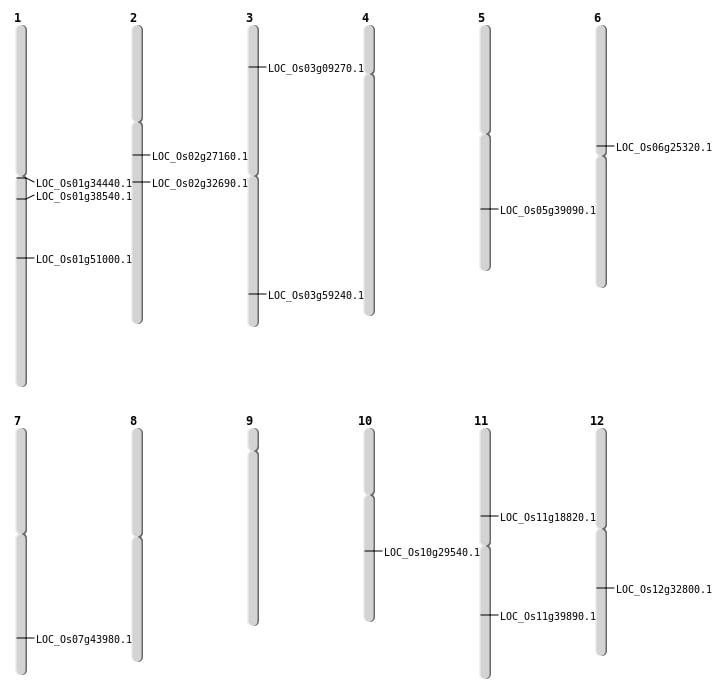
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12275472 | A-C | HET | INTERGENIC | |
| SBS | Chr1 | 16548889 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 18493893 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 18990612 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g34440 |
| SBS | Chr1 | 28353565 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 11895728 | A-C | HET | INTRON | |
| SBS | Chr10 | 473256 | T-A | HET | INTERGENIC | |
| SBS | Chr11 | 23776983 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g39890 |
| SBS | Chr11 | 6427084 | A-G | HET | INTERGENIC | |
| SBS | Chr12 | 10345270 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 25018053 | G-T | HOMO | INTRON | |
| SBS | Chr2 | 34921761 | T-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 34921762 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 34921763 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 3962439 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 10205474 | T-A | HET | INTRON | |
| SBS | Chr3 | 11171665 | T-C | HET | UTR_5_PRIME | |
| SBS | Chr3 | 13559330 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 19650989 | G-C | HET | INTERGENIC | |
| SBS | Chr3 | 27135667 | G-A | HET | INTRON | |
| SBS | Chr3 | 6282586 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 10929263 | T-A | HET | INTRON | |
| SBS | Chr4 | 17052098 | G-C | HET | SPLICE_SITE_REGION | |
| SBS | Chr4 | 25228149 | G-A | HET | INTRON | |
| SBS | Chr4 | 29110317 | G-A | HOMO | UTR_5_PRIME | |
| SBS | Chr4 | 3002231 | G-C | HET | INTRON | |
| SBS | Chr5 | 1554956 | T-G | HOMO | INTRON | |
| SBS | Chr5 | 3402769 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 19460993 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 812527 | A-G | HET | INTRON | |
| SBS | Chr7 | 19501180 | C-T | HOMO | INTRON | |
| SBS | Chr7 | 20923497 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 25715324 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 25715325 | A-T | HOMO | INTERGENIC | |
| SBS | Chr9 | 8808696 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 9232782 | G-A | HET | INTERGENIC |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 920051 | 920052 | 1 | |
| Deletion | Chr12 | 1505851 | 1505853 | 2 | |
| Deletion | Chr3 | 4844252 | 4844253 | 1 | LOC_Os03g09270 |
| Deletion | Chr9 | 8276605 | 8276611 | 6 | |
| Deletion | Chr10 | 9968930 | 9968932 | 2 | |
| Deletion | Chr1 | 10300321 | 10300338 | 17 | |
| Deletion | Chr6 | 10587535 | 10587547 | 12 | |
| Deletion | Chr11 | 10680843 | 10680844 | 1 | LOC_Os11g18820 |
| Deletion | Chr4 | 13472673 | 13472692 | 19 | |
| Deletion | Chr8 | 14307866 | 14307867 | 1 | |
| Deletion | Chr8 | 15005153 | 15005183 | 30 | |
| Deletion | Chr10 | 15342346 | 15342354 | 8 | LOC_Os10g29540 |
| Deletion | Chr2 | 15990106 | 15990107 | 1 | LOC_Os02g27160 |
| Deletion | Chr12 | 15998167 | 15998186 | 19 | |
| Deletion | Chr3 | 16135271 | 16135276 | 5 | |
| Deletion | Chr4 | 21301820 | 21301829 | 9 | |
| Deletion | Chr6 | 22274438 | 22274453 | 15 | |
| Deletion | Chr10 | 22533696 | 22533699 | 3 | |
| Deletion | Chr5 | 22922413 | 22922780 | 367 | |
| Deletion | Chr12 | 24047637 | 24047638 | 1 | |
| Deletion | Chr8 | 24104527 | 24104535 | 8 | |
| Deletion | Chr7 | 24983479 | 24983499 | 20 | |
| Deletion | Chr12 | 25817880 | 25817885 | 5 | |
| Deletion | Chr1 | 27208429 | 27208430 | 1 | |
| Deletion | Chr7 | 29010400 | 29010410 | 10 | |
| Deletion | Chr1 | 29302420 | 29302421 | 1 | LOC_Os01g51000 |
| Deletion | Chr3 | 29445743 | 29445757 | 14 | |
| Deletion | Chr3 | 33732590 | 33732606 | 16 | LOC_Os03g59240 |
| Deletion | Chr1 | 34235835 | 34235848 | 13 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 41514524 | 41514524 | 1 | |
| Insertion | Chr3 | 27345028 | 27345029 | 2 | |
| Insertion | Chr5 | 28034317 | 28034317 | 1 |
No Inversion
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 9527578 | Chr6 | 25514505 | |
| Translocation | Chr12 | 19818828 | Chr6 | 14832851 | 2 |
| Translocation | Chr5 | 22922414 | Chr1 | 21645754 | 2 |
| Translocation | Chr5 | 22922775 | Chr1 | 22260466 | LOC_Os05g39090 |
| Translocation | Chr7 | 26294740 | Chr2 | 19408949 | 2 |
| Translocation | Chr3 | 32459730 | Chr1 | 35462527 |
