Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN718-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN718-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 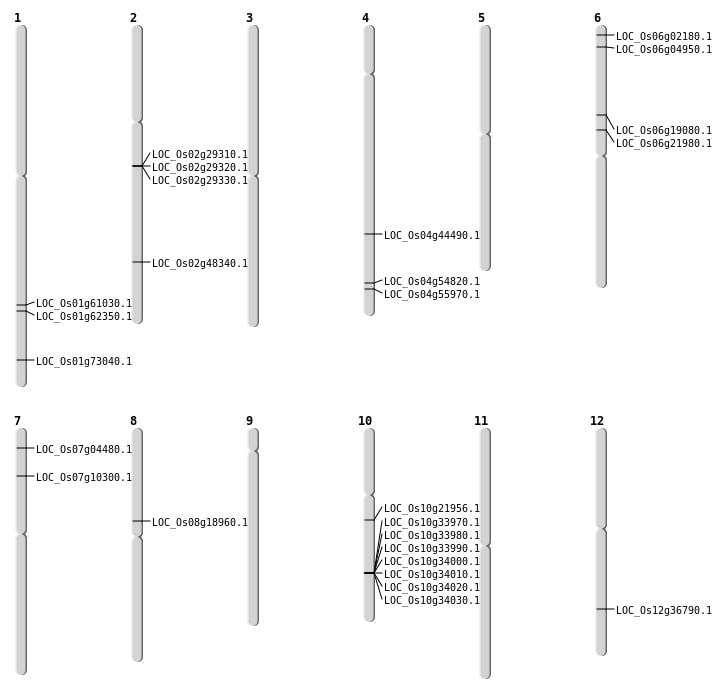
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 34019110 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 384086 | T-A | HET | INTRON | |
| SBS | Chr1 | 384087 | G-T | HET | INTRON | |
| SBS | Chr1 | 39465053 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 6964963 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 6964964 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 11369718 | G-A | HET | INTRON | |
| SBS | Chr10 | 13071906 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 17780339 | A-C | HET | INTRON | |
| SBS | Chr11 | 18056681 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 22555269 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g36790 |
| SBS | Chr2 | 17347490 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 17347491 | G-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 24972938 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 3429303 | C-T | HOMO | INTRON | |
| SBS | Chr3 | 1080996 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 12786431 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 28406547 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 30722267 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 22509716 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 26334631 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g44490 |
| SBS | Chr5 | 14169283 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 15296715 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 8211840 | T-A | HET | INTRON | |
| SBS | Chr6 | 11398885 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 25596569 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 20842447 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 9177270 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 21750557 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 24149962 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 6487435 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 17489993 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 22823835 | G-T | HET | INTERGENIC |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 1980122 | 1980127 | 5 | LOC_Os07g04480 |
| Deletion | Chr7 | 2656825 | 2656826 | 1 | |
| Deletion | Chr7 | 3813877 | 3813880 | 3 | |
| Deletion | Chr7 | 5526395 | 5526403 | 8 | LOC_Os07g10300 |
| Deletion | Chr1 | 5605079 | 5605082 | 3 | |
| Deletion | Chr9 | 7314134 | 7314136 | 2 | |
| Deletion | Chr1 | 7349248 | 7349273 | 25 | |
| Deletion | Chr9 | 8612897 | 8612900 | 3 | |
| Deletion | Chr8 | 9739000 | 9739019 | 19 | |
| Deletion | Chr8 | 11307430 | 11307501 | 71 | LOC_Os08g18960 |
| Deletion | Chr10 | 11530739 | 11530745 | 6 | |
| Deletion | Chr10 | 12349267 | 12349275 | 8 | |
| Deletion | Chr6 | 12707872 | 12716734 | 8862 | LOC_Os06g21980 |
| Deletion | Chr1 | 15089951 | 15089953 | 2 | |
| Deletion | Chr7 | 15271656 | 15271658 | 2 | |
| Deletion | Chr2 | 17412353 | 17426123 | 13770 | 3 |
| Deletion | Chr7 | 17825918 | 17825925 | 7 | |
| Deletion | Chr10 | 18100500 | 18153470 | 52970 | 7 |
| Deletion | Chr2 | 19937508 | 19937515 | 7 | |
| Deletion | Chr4 | 20281994 | 20282003 | 9 | |
| Deletion | Chr4 | 20898537 | 20898544 | 7 | |
| Deletion | Chr2 | 22459062 | 22459063 | 1 | |
| Deletion | Chr5 | 23816534 | 23816539 | 5 | |
| Deletion | Chr8 | 24273001 | 24273015 | 14 | |
| Deletion | Chr11 | 29004999 | 29005000 | 1 | |
| Deletion | Chr2 | 29598703 | 29598712 | 9 | LOC_Os02g48340 |
| Deletion | Chr3 | 29903593 | 29903616 | 23 | |
| Deletion | Chr4 | 32600754 | 32600755 | 1 | LOC_Os04g54820 |
| Deletion | Chr4 | 33343086 | 33345504 | 2418 | LOC_Os04g55970 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr6 | 27553767 | 27553767 | 1 | |
| Insertion | Chr6 | 700093 | 700094 | 2 | |
| Insertion | Chr8 | 143515 | 143516 | 2 | |
| Insertion | Chr9 | 15014805 | 15014808 | 4 |
Inversions: 9
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 661409 | 2180889 | 2 |
| Inversion | Chr6 | 661420 | 2180892 | 2 |
| Inversion | Chr1 | 6809694 | 7469284 | |
| Inversion | Chr1 | 6809716 | 7469311 | |
| Inversion | Chr4 | 22215103 | 33345506 | LOC_Os04g55970 |
| Inversion | Chr1 | 35315022 | 36084814 | 2 |
| Inversion | Chr1 | 35315033 | 36084814 | 2 |
| Inversion | Chr1 | 35562011 | 43023926 | |
| Inversion | Chr1 | 42381106 | 43023914 | LOC_Os01g73040 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 7641698 | Chr1 | 26359238 | |
| Translocation | Chr10 | 11338826 | Chr6 | 10850161 | 2 |
