Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN723-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN723-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 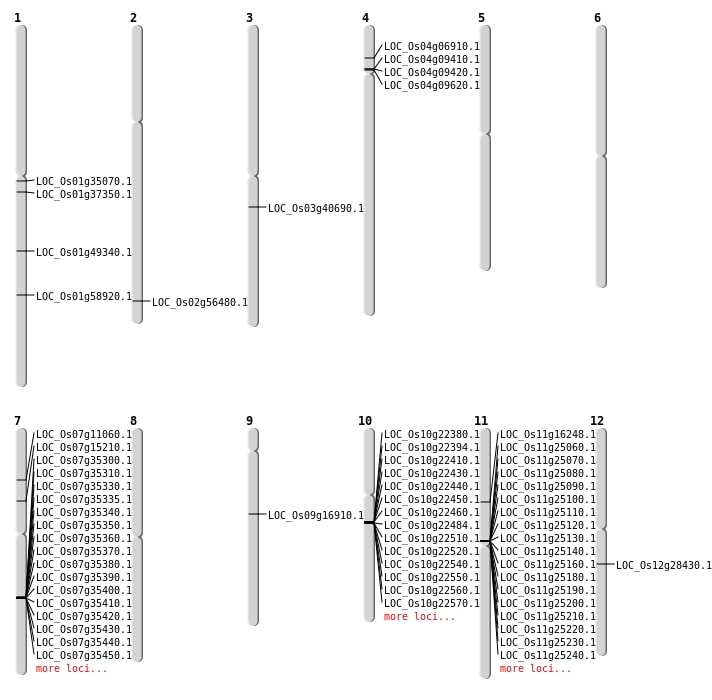
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18249609 | G-A | HOMO | INTRON | |
| SBS | Chr1 | 19419318 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g35070 |
| SBS | Chr1 | 20842511 | T-C | HOMO | UTR_5_PRIME | |
| SBS | Chr1 | 25245073 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 30057106 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 34055245 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g58920 |
| SBS | Chr1 | 34055246 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g58920 |
| SBS | Chr1 | 42540470 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 12037228 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 23067577 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g42754 |
| SBS | Chr10 | 23187781 | C-T | HOMO | INTRON | |
| SBS | Chr11 | 1480409 | G-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 18192471 | G-A | HET | STOP_GAINED | LOC_Os11g31210 |
| SBS | Chr11 | 18192472 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g31210 |
| SBS | Chr12 | 11157391 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 34575480 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os02g56480 |
| SBS | Chr2 | 4402124 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 7411015 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 20008960 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 22506014 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 22562328 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 30767250 | C-T | HET | INTRON | |
| SBS | Chr4 | 14966498 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 31431238 | C-T | HOMO | INTERGENIC | |
| SBS | Chr4 | 8908813 | G-C | HOMO | INTERGENIC | |
| SBS | Chr5 | 5536791 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 17057473 | C-G | HOMO | INTRON | |
| SBS | Chr6 | 17883718 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 1937764 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 8994596 | T-C | HOMO | INTRON | |
| SBS | Chr7 | 9091392 | T-C | HOMO | INTRON | |
| SBS | Chr8 | 6635764 | C-G | HET | INTRON | |
| SBS | Chr9 | 15024653 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 999299 | G-A | HET | INTERGENIC |
Deletions: 28
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 1728793 | 1728794 | 1 | |
| Deletion | Chr5 | 2674108 | 2674111 | 3 | |
| Deletion | Chr4 | 3649927 | 3649929 | 2 | LOC_Os04g06910 |
| Deletion | Chr4 | 5017001 | 5037000 | 20000 | 2 |
| Deletion | Chr4 | 5170142 | 5170146 | 4 | LOC_Os04g09620 |
| Deletion | Chr2 | 7151611 | 7151613 | 2 | |
| Deletion | Chr9 | 10333328 | 10333822 | 494 | LOC_Os09g16910 |
| Deletion | Chr10 | 11580264 | 11779321 | 199057 | 23 |
| Deletion | Chr11 | 14276844 | 14756383 | 479539 | 73 |
| Deletion | Chr1 | 14636421 | 14636430 | 9 | |
| Deletion | Chr9 | 14645905 | 14645942 | 37 | |
| Deletion | Chr10 | 14729267 | 14729268 | 1 | LOC_Os10g28310 |
| Deletion | Chr3 | 15363873 | 15363874 | 1 | |
| Deletion | Chr5 | 16501254 | 16501256 | 2 | |
| Deletion | Chr12 | 16807429 | 16807531 | 102 | LOC_Os12g28430 |
| Deletion | Chr6 | 16844183 | 16844184 | 1 | |
| Deletion | Chr10 | 16850001 | 17054000 | 204000 | 32 |
| Deletion | Chr6 | 17092460 | 17093576 | 1116 | |
| Deletion | Chr10 | 17326690 | 17756725 | 430035 | 54 |
| Deletion | Chr1 | 19418323 | 19418336 | 13 | LOC_Os01g35070 |
| Deletion | Chr2 | 20165100 | 20165103 | 3 | |
| Deletion | Chr1 | 20856905 | 20856910 | 5 | LOC_Os01g37350 |
| Deletion | Chr7 | 21127001 | 21249000 | 122000 | 23 |
| Deletion | Chr1 | 24102290 | 24102292 | 2 | |
| Deletion | Chr3 | 25605789 | 25605797 | 8 | |
| Deletion | Chr7 | 27974913 | 27974920 | 7 | |
| Deletion | Chr1 | 28370114 | 28370115 | 1 | LOC_Os01g49340 |
| Deletion | Chr3 | 36136166 | 36136167 | 1 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr12 | 2894463 | 2894463 | 1 | |
| Insertion | Chr4 | 22848265 | 22848266 | 2 | |
| Insertion | Chr5 | 25396395 | 25396395 | 1 | |
| Insertion | Chr7 | 8766150 | 8766150 | 1 | LOC_Os07g15210 |
| Insertion | Chr9 | 19168294 | 19168295 | 2 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 14754972 | 14756424 |
Translocations: 11
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 545945 | Chr2 | 26803932 | |
| Translocation | Chr11 | 548930 | Chr2 | 26803887 | |
| Translocation | Chr7 | 6114820 | Chr3 | 22628228 | 2 |
| Translocation | Chr7 | 6115289 | Chr3 | 22628258 | 2 |
| Translocation | Chr11 | 8939851 | Chr8 | 16045133 | LOC_Os11g16248 |
| Translocation | Chr9 | 10333329 | Chr7 | 5906568 | LOC_Os09g16910 |
| Translocation | Chr11 | 14276862 | Chr10 | 4992678 | |
| Translocation | Chr11 | 14754881 | Chr10 | 4992719 | |
| Translocation | Chr10 | 22706509 | Chr7 | 5906867 | LOC_Os10g42196 |
| Translocation | Chr10 | 22706521 | Chr9 | 10333869 | LOC_Os10g42196 |
| Translocation | Chr10 | 22706551 | Chr9 | 10333326 | 2 |
