Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN728-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN728-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 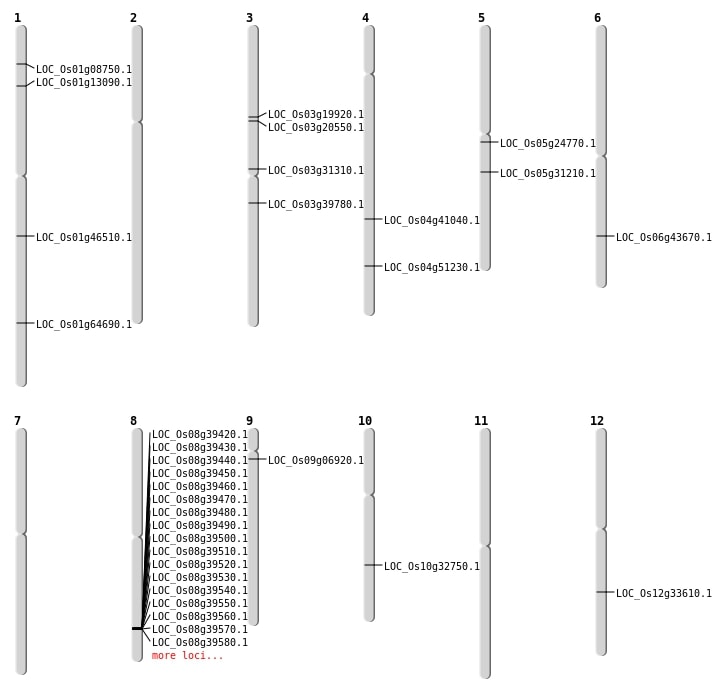
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 22511530 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 25151918 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 32933273 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 37528988 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g64690 |
| SBS | Chr1 | 3963833 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 3963834 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 3963836 | G-C | HET | INTERGENIC | |
| SBS | Chr10 | 17154489 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g32750 |
| SBS | Chr10 | 18694206 | C-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 5113859 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 1292332 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr11 | 1292333 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr11 | 20643021 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 16655006 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 2001816 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 20294507 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g33610 |
| SBS | Chr12 | 4514966 | A-T | HET | INTERGENIC | |
| SBS | Chr12 | 9423665 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 21908965 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 22684523 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 17844202 | A-C | HOMO | UTR_5_PRIME | |
| SBS | Chr3 | 19952129 | C-T | HET | INTRON | |
| SBS | Chr3 | 2471761 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 28239725 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 30641217 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 20785481 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 22621134 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 25080659 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 11120048 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 11120049 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 12921422 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 26825887 | T-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 7175638 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 2690504 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 27345491 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 27943503 | A-T | HET | INTRON | |
| SBS | Chr8 | 7846910 | A-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 8030686 | C-T | HOMO | INTERGENIC |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 3341402 | 3341403 | 1 | LOC_Os09g06920 |
| Deletion | Chr7 | 3973521 | 3973524 | 3 | |
| Deletion | Chr1 | 4376852 | 4376857 | 5 | LOC_Os01g08750 |
| Deletion | Chr1 | 7283707 | 7283728 | 21 | LOC_Os01g13090 |
| Deletion | Chr9 | 8256903 | 8256906 | 3 | |
| Deletion | Chr11 | 8524916 | 8524917 | 1 | |
| Deletion | Chr3 | 11202011 | 11202022 | 11 | LOC_Os03g19920 |
| Deletion | Chr3 | 11650973 | 11650978 | 5 | LOC_Os03g20550 |
| Deletion | Chr5 | 14359356 | 14359358 | 2 | LOC_Os05g24770 |
| Deletion | Chr3 | 17844203 | 17844205 | 2 | LOC_Os03g31310 |
| Deletion | Chr8 | 17918653 | 17918662 | 9 | |
| Deletion | Chr5 | 18133356 | 18133379 | 23 | |
| Deletion | Chr5 | 18150636 | 18150637 | 1 | LOC_Os05g31210 |
| Deletion | Chr12 | 19448997 | 19448998 | 1 | |
| Deletion | Chr8 | 20356736 | 20356748 | 12 | |
| Deletion | Chr3 | 22133365 | 22133366 | 1 | LOC_Os03g39780 |
| Deletion | Chr10 | 22209607 | 22209608 | 1 | |
| Deletion | Chr4 | 24353284 | 24353286 | 2 | LOC_Os04g41040 |
| Deletion | Chr8 | 24924001 | 25075000 | 151000 | 21 |
| Deletion | Chr4 | 25071557 | 25071558 | 1 | |
| Deletion | Chr5 | 25750033 | 25750034 | 1 | |
| Deletion | Chr6 | 26280067 | 26280074 | 7 | LOC_Os06g43670 |
| Deletion | Chr1 | 26454508 | 26454637 | 129 | LOC_Os01g46510 |
| Deletion | Chr8 | 27218508 | 27218509 | 1 | |
| Deletion | Chr4 | 30352622 | 30352623 | 1 | LOC_Os04g51230 |
| Deletion | Chr2 | 30488181 | 30488185 | 4 | |
| Deletion | Chr4 | 35302903 | 35302904 | 1 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr4 | 25687811 | 25687812 | 2 | |
| Insertion | Chr6 | 13454925 | 13454927 | 3 | |
| Insertion | Chr6 | 9763365 | 9763365 | 1 |
No Inversion
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 9018068 | Chr8 | 21985176 | |
| Translocation | Chr11 | 19817759 | Chr1 | 39521938 |
