Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN739-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN739-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 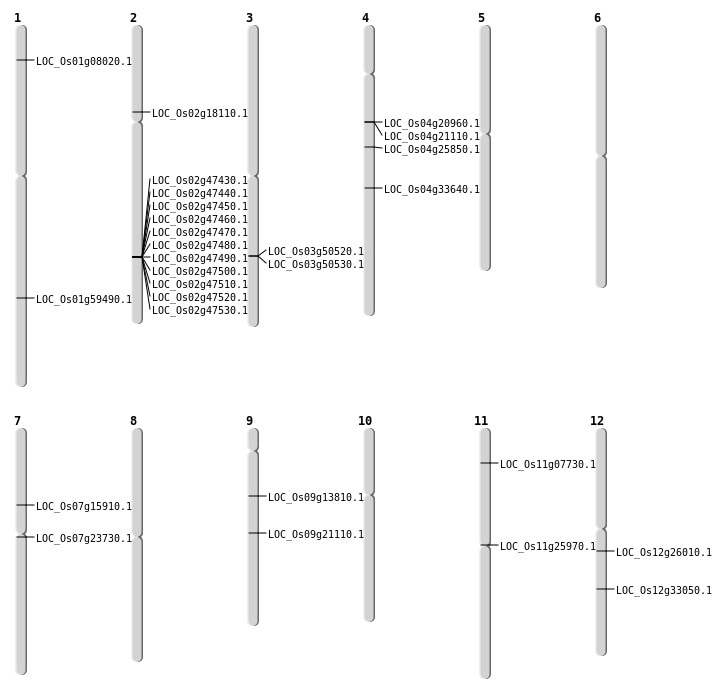
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14676247 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 1810029 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 22682908 | C-G | HET | INTERGENIC | |
| SBS | Chr1 | 22865359 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 10595825 | A-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 20259063 | A-T | HOMO | UTR_3_PRIME | |
| SBS | Chr10 | 9009080 | C-T | HOMO | INTRON | |
| SBS | Chr11 | 1117111 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 14516679 | A-G | HOMO | INTERGENIC | |
| SBS | Chr11 | 14516681 | T-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 14516682 | C-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 26850045 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 27587669 | G-A | HET | INTRON | |
| SBS | Chr11 | 3947268 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g07730 |
| SBS | Chr12 | 11800489 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 14970991 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 15111814 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g26010 |
| SBS | Chr2 | 27830085 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 17283786 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 2187377 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 23721164 | A-T | HET | INTRON | |
| SBS | Chr4 | 12082710 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 20376166 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g33640 |
| SBS | Chr4 | 34499168 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr4 | 8323878 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 26417080 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 6799587 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 24690139 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 25913937 | G-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 8480715 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 13111591 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 14221291 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 15102752 | G-A | HET | INTRON | |
| SBS | Chr8 | 22509271 | T-G | HET | INTRON | |
| SBS | Chr8 | 2275999 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 11245517 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 1206636 | G-A | HET | INTRON | |
| SBS | Chr9 | 13063837 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 7793930 | T-C | HET | INTERGENIC |
Deletions: 23
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 1739739 | 1739740 | 1 | |
| Deletion | Chr1 | 3878115 | 3878125 | 10 | LOC_Os01g08020 |
| Deletion | Chr5 | 6799580 | 6799581 | 1 | |
| Deletion | Chr3 | 7546989 | 7546993 | 4 | |
| Deletion | Chr6 | 8423009 | 8423010 | 1 | |
| Deletion | Chr7 | 9247159 | 9247177 | 18 | LOC_Os07g15910 |
| Deletion | Chr11 | 10749632 | 10749636 | 4 | |
| Deletion | Chr7 | 13391386 | 13391393 | 7 | LOC_Os07g23730 |
| Deletion | Chr11 | 14808001 | 14819000 | 11000 | LOC_Os11g25970 |
| Deletion | Chr6 | 16354783 | 16354785 | 2 | |
| Deletion | Chr2 | 18070671 | 18070677 | 6 | |
| Deletion | Chr12 | 19973545 | 19973549 | 4 | LOC_Os12g33050 |
| Deletion | Chr12 | 23867099 | 23867100 | 1 | |
| Deletion | Chr5 | 27646289 | 27646292 | 3 | |
| Deletion | Chr7 | 27995332 | 27995339 | 7 | |
| Deletion | Chr3 | 28852215 | 28852228 | 13 | 2 |
| Deletion | Chr2 | 28961001 | 29041000 | 80000 | 11 |
| Deletion | Chr4 | 31452567 | 31452579 | 12 | |
| Deletion | Chr1 | 34409129 | 34409137 | 8 | LOC_Os01g59490 |
| Deletion | Chr4 | 34920405 | 34920428 | 23 | |
| Deletion | Chr1 | 36360835 | 36360847 | 12 | |
| Deletion | Chr1 | 36405661 | 36405675 | 14 | |
| Deletion | Chr1 | 36474219 | 36474221 | 2 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr3 | 181913 | 181914 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 12668769 | 12734647 | LOC_Os09g21110 |
| Inversion | Chr5 | 12742354 | 12818803 | |
| Inversion | Chr11 | 14807659 | 19233110 | |
| Inversion | Chr11 | 14818627 | 19232373 | LOC_Os11g25970 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 6993245 | Chr3 | 22046068 | |
| Translocation | Chr9 | 8102895 | Chr4 | 11882668 | 2 |
| Translocation | Chr9 | 8102904 | Chr4 | 11780229 | 2 |
| Translocation | Chr4 | 15030926 | Chr2 | 10522453 | 2 |
