Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN747-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN747-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 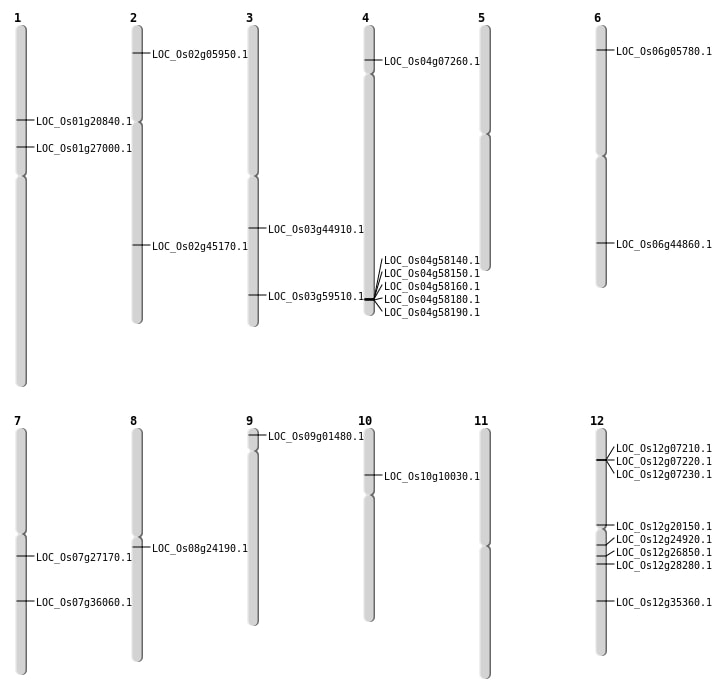
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15043653 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g27000 |
| SBS | Chr1 | 18933940 | G-A | HET | INTRON | |
| SBS | Chr1 | 20631203 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 35731286 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 5917413 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 12300840 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 19502870 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 4603621 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 4638005 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 10796924 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 11893442 | G-A | HET | INTRON | |
| SBS | Chr12 | 14762811 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 2306675 | A-T | HET | SPLICE_SITE_REGION | |
| SBS | Chr12 | 23267005 | G-A | HET | INTRON | |
| SBS | Chr2 | 17098381 | A-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 24036454 | G-A | HOMO | INTRON | |
| SBS | Chr2 | 30630972 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 17648050 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 28737210 | G-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 29219443 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 33872152 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g59510 |
| SBS | Chr3 | 7195866 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr5 | 14395523 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 20896796 | G-A | HET | INTRON | |
| SBS | Chr5 | 23964837 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr6 | 13451574 | C-T | HET | INTRON | |
| SBS | Chr6 | 19754419 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 2622009 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g05780 |
| SBS | Chr6 | 5643951 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 21565052 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g36060 |
| SBS | Chr8 | 12761084 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 14007570 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 9009960 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 351587 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g01480 |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 2947384 | 2947385 | 1 | |
| Deletion | Chr2 | 2961726 | 2961735 | 9 | LOC_Os02g05950 |
| Deletion | Chr11 | 3123681 | 3123685 | 4 | |
| Deletion | Chr12 | 3540001 | 3555000 | 15000 | 3 |
| Deletion | Chr12 | 4604616 | 4604622 | 6 | |
| Deletion | Chr10 | 5448000 | 5448027 | 27 | LOC_Os10g10030 |
| Deletion | Chr8 | 6512817 | 6512822 | 5 | |
| Deletion | Chr10 | 6814663 | 6814667 | 4 | |
| Deletion | Chr8 | 8238915 | 8238931 | 16 | |
| Deletion | Chr11 | 8996797 | 8996798 | 1 | |
| Deletion | Chr2 | 11469482 | 11469493 | 11 | |
| Deletion | Chr12 | 11729014 | 11735077 | 6063 | LOC_Os12g20150 |
| Deletion | Chr2 | 15788095 | 15788096 | 1 | |
| Deletion | Chr3 | 16410504 | 16410513 | 9 | |
| Deletion | Chr5 | 17377734 | 17377777 | 43 | |
| Deletion | Chr6 | 18121443 | 18121444 | 1 | |
| Deletion | Chr7 | 19431321 | 19431332 | 11 | |
| Deletion | Chr8 | 20887098 | 20887099 | 1 | |
| Deletion | Chr1 | 23071552 | 23071570 | 18 | |
| Deletion | Chr7 | 23743000 | 23743005 | 5 | |
| Deletion | Chr3 | 25355963 | 25355965 | 2 | LOC_Os03g44910 |
| Deletion | Chr6 | 26827552 | 26827555 | 3 | |
| Deletion | Chr6 | 27110660 | 27115006 | 4346 | LOC_Os06g44860 |
| Deletion | Chr2 | 27419786 | 27419797 | 11 | LOC_Os02g45170 |
| Deletion | Chr4 | 28304272 | 28304277 | 5 | |
| Deletion | Chr4 | 34625766 | 34655665 | 29899 | 5 |
Insertions: 6
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 11080603 | 11207705 | |
| Inversion | Chr8 | 13290677 | 14601578 | LOC_Os08g24190 |
| Inversion | Chr8 | 13290822 | 14601585 | LOC_Os08g24190 |
| Inversion | Chr12 | 14313174 | 17067164 | LOC_Os12g24920 |
| Inversion | Chr12 | 15729143 | 16722226 | 2 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 5829886 | Chr1 | 11618352 | LOC_Os01g20840 |
| Translocation | Chr7 | 15760090 | Chr4 | 3855204 | 2 |
| Translocation | Chr12 | 21494739 | Chr11 | 17511104 | LOC_Os12g35360 |
| Translocation | Chr11 | 25331002 | Chr2 | 2430960 |
